Identification of a Trm732 Motif Required for 2'- O-methylation of the tRNA Anticodon Loop by Trm7
- PMID: 35559166
- PMCID: PMC9088939
- DOI: 10.1021/acsomega.1c07231
Identification of a Trm732 Motif Required for 2'- O-methylation of the tRNA Anticodon Loop by Trm7
Abstract
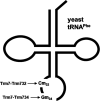
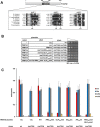
Posttranscriptional tRNA modifications are essential for proper gene expression, and defects in the enzymes that perform tRNA modifications are associated with numerous human disorders. Throughout eukaryotes, 2'-O-methylation of residues 32 and 34 of the anticodon loop of tRNA is important for proper translation, and in humans, a lack of these modifications results in non-syndromic X-linked intellectual disability. In yeast, the methyltransferase Trm7 forms a complex with Trm732 to 2'-O-methylate tRNA residue 32 and with Trm734 to 2'-O-methylate tRNA residue 34. Trm732 and Trm734 are required for the methylation activity of Trm7, but the role of these auxiliary proteins is not clear. Additionally, Trm732 and Trm734 homologs are implicated in biological processes not directly related to translation, suggesting that these proteins may have additional cellular functions. To identify critical amino acids in Trm732, we generated variants and tested their ability to function in yeast cells. We identified a conserved RRSAGLP motif in the conserved DUF2428 domain of Trm732 that is required for tRNA modification activity by both yeast Trm732 and its human homolog, THADA. The identification of Trm732 variants that lack tRNA modification activity will help to determine if other biological functions ascribed to Trm732 and THADA are directly due to tRNA modification or to secondary effects due to other functions of these proteins.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Boccaletto P.; Machnicka M. A.; Purta E.; Piątkowski P.; Bagiński B.; Wirecki T. K.; de Crécy-Lagard V.; Ross R.; Limbach P. A.; Kotter A.; Helm M.; Bujnicki J. M. MODOMICS: A Database of RNA Modification Pathways. 2017 Update. Nucleic Acids Res. 2018, 46, D303–D307. 10.1093/nar/gkx1030. - DOI - PMC - PubMed
-
- Cuajungco M. P.; Leyne M.; Mull J.; Gill S. P.; Lu W.; Zagzag D.; Axelrod F. B.; Maayan C.; Gusella J. F.; Slaugenhaupt S. A. Tissue-Specific Reduction in Splicing Efficiency of IKBKAP Due to the Major Mutation Associated with Familial Dysautonomia. Am. J. Hum. Genet. 2003, 72, 749–758. 10.1086/368263. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
