CRISPR/Cas9-mediated tetra-allelic mutation of the 'Green Revolution' SEMIDWARF-1 (SD-1) gene confers lodging resistance in tef (Eragrostis tef)
- PMID: 35560779
- PMCID: PMC9398311
- DOI: 10.1111/pbi.13842
CRISPR/Cas9-mediated tetra-allelic mutation of the 'Green Revolution' SEMIDWARF-1 (SD-1) gene confers lodging resistance in tef (Eragrostis tef)
Abstract
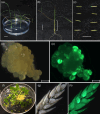
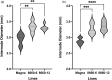
Tef is a staple food and a valuable cash crop for millions of people in Ethiopia. Lodging is a major limitation to tef production, and for decades, the development of lodging resistant varieties proved difficult with conventional breeding approaches. We used CRISPR/Cas9 to introduce knockout mutations in the tef orthologue of the rice SEMIDWARF-1 (SD-1) gene to confer semidwarfism and ultimately lodging resistance. High frequency recovery of transgenic and SD-1 edited tef lines was achieved in two tef cultivars by Agrobacterium-mediated delivery into young leaf explants of gene editing reagents along with transformation and regeneration enhancing morphogenic genes, BABY BOOM (BBM) and WUSCHEL2 (WUS2). All of the 23 lines analyzed by next-generation sequencing had at least two or more alleles of SD-1 mutated. Of these, 83% had tetra-allelic frameshift mutations in the SD-1 gene in primary tef regenerants, which were inherited in subsequent generations. Phenotypic data generated on T1 and T2 generations revealed that the sd-1 lines have reduced culm and internode lengths with no reduction in either panicle or peduncle lengths. These characteristics are comparable with rice sd-1 plants. Measurements of lodging, in greenhouse-grown plants, showed that sd-1 lines have significantly higher resistance to lodging at the heading stage compared with the controls. This is the first demonstration of the feasibility of high frequency genetic transformation and CRISPR/Cas9-mediated genome editing in this highly valuable but neglected crop. The findings reported here highlight the potential of genome editing for the improvement of lodging resistance and other important traits in tef.
Keywords: BABY BOOM; Eragrostis tef; SEMIDWARF-1; WUSCHEL; CRISPR/Cas9; lodging.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Achard, P. and Genschik, P. (2009) Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 60, 1085–1092. - PubMed
-
- Ashikari, M. , Sasaki, A. , Ueguchi‐Tanaka, M. , Itoh, H. , Nishimura, A. , Datta, S. , Ishiyama, K. et al. (2002) Loss‐of‐function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox‐2), led to the rice ‘green revolution’. Breed. Sci. 52, 143–150.
-
- Assefa, K. and Chanyalew, S. (2018) Agronomics of tef. In Minten, B., Taffesse, A.S. and Brown, P. eds., The Economics of Tef, Exploring Ethiopia’s Biggest Cash Crop, pp. 39–70. Wasington, DC: International Food Policy Research Institute (IFPRI).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

