Metabolic reprogramming of the intestinal microbiome with functional bile acid changes underlie the development of NAFLD
- PMID: 35561146
- PMCID: PMC9653520
- DOI: 10.1002/hep.32568
Metabolic reprogramming of the intestinal microbiome with functional bile acid changes underlie the development of NAFLD
Abstract
Background and aims: Bile acids are hepatic metabolites and have many properties considered to be relevant to the pathophysiology of NAFLD. Circulating levels of the intestinal microbiome-modified bile acid deoxycholate are increased in cirrhosis.
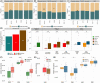
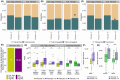
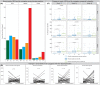
Approach and results: To further elucidate the role of bile acids and intestinal microbiota linked to bile acids in progressively severe NAFLD, a multiomic study of feces including 16S rRNA sequencing, microbial transcriptomics and metabolomics was performed in a cohort with varying phenotypes of NAFLD. Several bile acids of microbial origin derived from deoxycholic acid (DCA) (glycodeoxycholate, 7-ketodeoxycholic acid, dehydrocholic acid) increased with disease activity and fibrosis stage. These were linked to increased expression of microbial bile salt hydrolase, bile acid operon (BaiCD) and hydroxysteroid dehydrogenases (hdhA) required for DCA and downstream metabolite synthesis providing a mechanistic basis for altered bile acid profiles with disease progression. Bacteroidetes and several genera of Lachnospiraceae family containing DCA generating genes increased with increasing disease severity, whereas several potentially beneficial microbes sensitive to antibacterial effects of DCA e.g., Ruminococcaceae were decreased. The clinical relevance of these data was confirmed in an independent cohort enrolled in a clinical trial for NASH where at entry DCA and its conjugates were associated with advanced fibrosis. In patients treated with placebo, DCA declined in those with fibrosis regression and increased in those with fibrosis progression. DCA rose further in those with compensated cirrhosis when they experienced decompensation.
Conclusions: These findings demonstrate a role for bile acids and the bile acid dependent microbiome in the development and progression of NAFLD and set the stage to leverage these findings for NASH biomarker development and for therapeutics.
© 2022 The Authors. Hepatology published by Wiley Periodicals LLC on behalf of American Association for the Study of Liver Diseases.
Conflict of interest statement
Andrew N. Billin owns stock in and is employed by Gilead Sciences. Jen‐Chieh Chuang owns stock in and is employed by Gilead Sciences and The Liver Company. Ryan S. Huss owns stock in and is employed by The Liver Company. He owns stock in Gilead Sciences. Robert P. Myers owns stock in and is employed by The Liver Company. He owns stock in Gilead Sciences. Nicole Narayan owns stock in and is employed by Second Genome. Arun J. Sanyal consults and received grants from Conatus, Gilead, Malinckrodt, Immuron, Boehringer Ingelhiem, Novartis, Bristol Myers Squibb, Merck, Lilly, Novo Nordisk, Fractyl, Siemens, Madrigal, Inventiva, and Covance. He owns stock in and consults for Genfit and Hemoshear. He consults for Intercept, Pfizer, Salix, Galectin, Sequana, Terns, Albireo, Sanofi, Jannsen, Takeda, Northsea, AMRA, Perspectum, Poxel, 89 Bio, AstraZeneca, NGM Bio, Amgen, Regeneron, Genetech, Roche, Albireo, Prosciento, Histoindex, Path AI, and Biocellvia. He received grants from Echosense‐Sandhill, OWL, and Second Genome. He received royalties from Elsevier and UptoDate. He owns stock in Sanyal Bio, Exhalenz, Durect, Indalo, Tiziana, and Rivus.
Figures



References
-
- Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67(1):328–57. - PubMed
-
- Han MAT, Altayar O, Hamdeh S, Takyar V, Rotman Y, Etzion O, et al. Rates of and factors associated with placebo response in trials of pharmacotherapies for nonalcoholic steatohepatitis: systematic review and meta‐analysis. Clin Gastroenterol Hepatol. 2019;17(4):616–29.e26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

