Epigenetic Repression of STING by MYC Promotes Immune Evasion and Resistance to Immune Checkpoint Inhibitors in Triple-Negative Breast Cancer
- PMID: 35561311
- PMCID: PMC9250627
- DOI: 10.1158/2326-6066.CIR-21-0826
Epigenetic Repression of STING by MYC Promotes Immune Evasion and Resistance to Immune Checkpoint Inhibitors in Triple-Negative Breast Cancer
Abstract
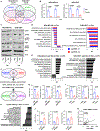
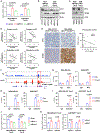
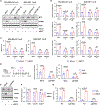
The MYC oncogene is frequently amplified in triple-negative breast cancer (TNBC). Here, we show that MYC suppression induces immune-related hallmark gene set expression and tumor-infiltrating T cells in MYC-hyperactivated TNBCs. Mechanistically, MYC repressed stimulator of interferon genes (STING) expression via direct binding to the STING1 enhancer region, resulting in downregulation of the T-cell chemokines CCL5, CXCL10, and CXCL11. In primary and metastatic TNBC cohorts, tumors with high MYC expression or activity exhibited low STING expression. Using a CRISPR-mediated enhancer perturbation approach, we demonstrated that MYC-driven immune evasion is mediated by STING repression. STING repression induced resistance to PD-L1 blockade in mouse models of TNBC. Finally, a small-molecule inhibitor of MYC combined with PD-L1 blockade elicited a durable response in immune-cold TNBC with high MYC expression, suggesting a strategy to restore PD-L1 inhibitor sensitivity in MYC-overexpressing TNBC.
©2022 American Association for Cancer Research.
Figures



References
-
- Schmid P, Rugo HS, Adams S, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2020;21(1):44–59 doi 10.1016/S1470-2045(19)30689-8. - DOI - PubMed
-
- Mittendorf EA, Zhang H, Barrios CH, Saji S, Jung KH, Hegg R, et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet 2020;396(10257):1090–100 doi 10.1016/S0140-6736(20)31953-X. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

