Whole-genome sequencing of Mycobacterium tuberculosis from Cambodia
- PMID: 35562174
- PMCID: PMC9095694
- DOI: 10.1038/s41598-022-10964-9
Whole-genome sequencing of Mycobacterium tuberculosis from Cambodia
Abstract
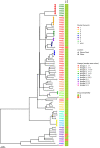
Cambodia has one of the highest tuberculosis (TB) incidence rates in the WHO Western Pacific region. Remarkably though, the prevalence of multidrug-resistant TB (MDR-TB) remains low. We explored the genetic diversity of Mycobacterium tuberculosis (MTB) circulating in this unique setting using whole-genome sequencing (WGS). From October 2017 until January 2018, we collected one hundred sputum specimens from consenting adults older than 21 years of age, newly diagnosed with bacteriologically confirmed TB in 3 districts of Phnom Penh and Takeo provinces of Cambodia before they commence on their TB treatment, where eighty MTB isolates were successfully cultured and sequenced. Majority of the isolates belonged to Lineage 1 (Indo-Oceanic) (69/80, 86.25%), followed by Lineage 2 (East Asian) (10/80, 12.5%) and Lineage 4 (Euro-American) (1/80, 1.25%). Phenotypic resistance to both streptomycin and isoniazid was found in 3 isolates (3/80, 3.75%), while mono-resistance to streptomycin and isoniazid was identical at 2.5% (N = 2 each). None of the isolates tested was resistant to either rifampicin or ethambutol. The specificities of genotypic prediction for resistance to all drugs tested were 100%, while the sensitivities of genotypic resistance predictions to isoniazid and streptomycin were lower at 40% (2/5) and 80% (4/5) respectively. We identified 8 clusters each comprising of two to five individuals all residing in the Takeo province, making up half (28/56, 50%) of all individuals sampled in the province, indicating the presence of multiple ongoing transmission events. All clustered isolates were of Lineage 1 and none are resistant to any of the drugs tested. This study while demonstrating the relevance and utility of WGS in predicting drug resistance and inference of disease transmission, highlights the need to increase the representation of genotype-phenotype TB data from low and middle income countries in Asia and Africa to improve the accuracies for prediction of drug resistance.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Organization, W.H. Global Tuberculosis Report 2018 (World Health Organization, 2018).
-
- Yamada N, et al. The national tuberculosis drug resistance survey in Cambodia, 2000–2001. Int. J. Tuberc. Lung Dis. 2007;11(12):1321–1327. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

