Rapid acceleration of KRAS-mutant pancreatic carcinogenesis via remodeling of tumor immune microenvironment by PPARδ
- PMID: 35562376
- PMCID: PMC9106716
- DOI: 10.1038/s41467-022-30392-7
Rapid acceleration of KRAS-mutant pancreatic carcinogenesis via remodeling of tumor immune microenvironment by PPARδ
Abstract
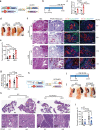
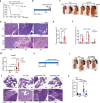
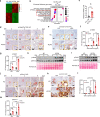
Pancreatic intraepithelial neoplasia (PanIN) is a precursor of pancreatic ductal adenocarcinoma (PDAC), which commonly occurs in the general populations with aging. Although most PanIN lesions (PanINs) harbor oncogenic KRAS mutations that initiate pancreatic tumorigenesis; PanINs rarely progress to PDAC. Critical factors that promote this progression, especially targetable ones, remain poorly defined. We show that peroxisome proliferator-activated receptor-delta (PPARδ), a lipid nuclear receptor, is upregulated in PanINs in humans and mice. Furthermore, PPARδ ligand activation by a high-fat diet or GW501516 (a highly selective, synthetic PPARδ ligand) in mutant KRASG12D (KRASmu) pancreatic epithelial cells strongly accelerates PanIN progression to PDAC. This PPARδ activation induces KRASmu pancreatic epithelial cells to secrete CCL2, which recruits immunosuppressive macrophages and myeloid-derived suppressor cells into pancreas via the CCL2/CCR2 axis to orchestrate an immunosuppressive tumor microenvironment and subsequently drive PanIN progression to PDAC. Our data identify PPARδ signaling as a potential molecular target to prevent PDAC development in subjects harboring PanINs.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Matsuda, Y. et al. The prevalence and clinicopathological characteristics of high-grade pancreatic intraepithelial neoplasia: autopsy study evaluating the entire pancreatic parenchyma. Pancreas46, 658–664 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

