Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance
- PMID: 35562425
- PMCID: PMC9099300
- DOI: 10.1038/s41580-022-00483-w
Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance
Abstract
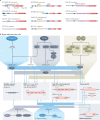
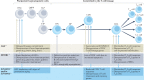
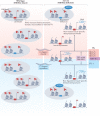
Heterochromatin is characterized by dimethylated or trimethylated histone H3 Lys9 (H3K9me2 or H3K9me3, respectively) and is found at transposable elements, satellite repeats and genes, where it ensures their transcriptional silencing. The histone methyltransferases (HMTs) that methylate H3K9 - in mammals Suppressor of variegation 3-9 homologue 1 (SUV39H1), SUV39H2, SET domain bifurcated 1 (SETDB1), SETDB2, G9A and G9A-like protein (GLP) - and the 'readers' of H3K9me2 or H3K9me3 are highly conserved and show considerable redundancy. Despite their redundancy, genetic ablation or mistargeting of an individual H3K9 methyltransferase can correlate with impaired cell differentiation, loss of tissue identity, premature aging and/or cancer. In this Review, we discuss recent advances in understanding the roles of the known H3K9-specific HMTs in ensuring transcriptional homeostasis during tissue differentiation in mammals. We examine the effects of H3K9-methylation-dependent gene repression in haematopoiesis, muscle differentiation and neurogenesis in mammals, and compare them with mechanistic insights obtained from the study of model organisms, notably Caenorhabditis elegans and Drosophila melanogaster. In all these organisms, H3K9-specific HMTs have both unique and redundant roles that ensure the maintenance of tissue integrity by restricting the binding of transcription factors to lineage-specific promoters and enhancer elements.
© 2022. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

