Discovery of Genomic Regions and Candidate Genes Controlling Root Development Using a Recombinant Inbred Line Population in Rapeseed (Brassica napus L.)
- PMID: 35563170
- PMCID: PMC9102059
- DOI: 10.3390/ijms23094781
Discovery of Genomic Regions and Candidate Genes Controlling Root Development Using a Recombinant Inbred Line Population in Rapeseed (Brassica napus L.)
Abstract
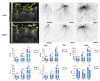
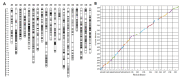
Marker-assisted selection enables breeders to quickly select excellent root architectural variations, which play an essential role in plant productivity. Here, ten root-related and shoot biomass traits of a new F6 recombinant inbred line (RIL) population were investigated under hydroponics and resulted in high heritabilities from 0.61 to 0.83. A high-density linkage map of the RIL population was constructed using a Brassica napus 50k Illumina single nucleotide polymorphism (SNP) array. A total of 86 quantitative trait loci (QTLs) explaining 4.16-14.1% of the phenotypic variances were detected and integrated into eight stable QTL clusters, which were repeatedly detected in different experiments. The codominant markers were developed to be tightly linked with three major QTL clusters, qcA09-2, qcC08-2, and qcC08-3, which controlled both root-related and shoot biomass traits and had phenotypic contributions greater than 10%. Among these, qcA09-2, renamed RT.A09, was further fine-mapped to a 129-kb interval with 19 annotated genes in the B. napus reference genome. By integrating the results of real-time PCR and comparative sequencing, five genes with expression differences and/or amino acid differences were identified as important candidate genes for RT.A09. Our findings laid the foundation for revealing the molecular mechanism of root development and developed valuable markers for root genetic improvement in rapeseed.
Keywords: fine mapping; linkage analysis; major QTL; rapeseed; root development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Grants and funding
- 31970526/National Natural Science Foundation of China
- 2020BBB061/Key Research and Development Program in Hubei Province
- CAAS-ZDRW202105/Agricultural Science and Technology Innovation Project
- CAAS-ASTIP-2013-OCRI/Agricultural Science and Technology Innovation Project
- CARS-12/China Agriculture Research System of MOF and MARA
LinkOut - more resources
Full Text Sources

