Long Non-Coding LEF1-AS1 Sponge miR-5100 Regulates Apoptosis and Autophagy in Gastric Cancer Cells via the miR-5100/DEK/AMPK-mTOR Axis
- PMID: 35563178
- PMCID: PMC9101949
- DOI: 10.3390/ijms23094787
Long Non-Coding LEF1-AS1 Sponge miR-5100 Regulates Apoptosis and Autophagy in Gastric Cancer Cells via the miR-5100/DEK/AMPK-mTOR Axis
Abstract
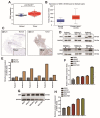
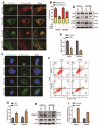
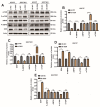
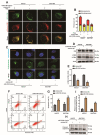
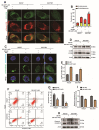
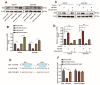
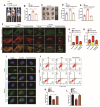
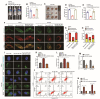
DEK and miR-5100 play critical roles in many steps of cancer initiation and progression and are directly or indirectly regulated by most promoters and repressors. LEF1-AS1 as a long non-coding RNA can regulate tumor development through sponge miRNA. The effect and regulatory mechanism of DEK on autophagy and apoptosis in gastric cancer (GC), and the role between miR-5100 and DEK or miR-5100 and LEF1-AS1 are still unclear. Our study found that DEK was highly expressed in gastric cancer tissues and cell lines, and knockdown of DEK inhibited the autophagy of cells, promoted apoptosis, and suppressed the malignant phenotype of gastric cancer. DEK regulates autophagy and apoptosis through the AMPK/mTOR signaling pathway. In addition, miR-5100 inhibits autophagy and promotes apoptosis in GC cells while LEF1-AS1 had the opposite effect. Studies have shown that miR-5100 acts by targeting the 3'UTR of DEK, and LEF1-AS1 regulates the expression of miR-5100 by sponging with mIR-5100. In conclusion, our results found that LEF1-AS1 and miR-5100 sponge function, and the miR-5100/DEK/AMPK/mTOR axis regulates autophagy and apoptosis in gastric cancer cells.
Keywords: DEK; LEF1-AS1; apoptosis; autophagy; miR-5100.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures


References
-
- Recio-Boiles A., Babiker H.M. StatPearls. StatPearls Publishing LLC; Treasure Island, FL, USA: 2022. Gastric Cancer. StatPearls Publishing Copyright © 2022.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

