Single-Cell RNA Sequencing Analysis for Oncogenic Mechanisms Underlying Oral Squamous Cell Carcinoma Carcinogenesis with Candida albicans Infection
- PMID: 35563222
- PMCID: PMC9104272
- DOI: 10.3390/ijms23094833
Single-Cell RNA Sequencing Analysis for Oncogenic Mechanisms Underlying Oral Squamous Cell Carcinoma Carcinogenesis with Candida albicans Infection
Abstract
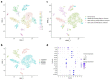
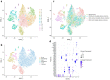
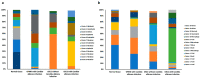
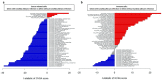
Oral squamous cell carcinoma (OSCC) carcinogenesis involves heterogeneous tumor cells, and the tumor microenvironment (TME) is highly complex with many different cell types. Cancer cell-TME interactions are crucial in OSCC progression. Candida albicans (C. albicans)-frequently pre-sent in the oral potentially malignant disorder (OPMD) lesions and OSCC tissues-promotes malignant transformation. The aim of the study is to verify the mechanisms underlying OSCC car-cinogenesis with C. albicans infection and identify the biomarker for the early detection of OSCC and as the treatment target. The single-cell RNA sequencing analysis (scRNA-seq) was performed to explore the cell subtypes in normal oral mucosa, OPMD, and OSCC tissues. The cell composi-tion changes and oncogenic mechanisms underlying OSCC carcinogenesis with C. albicans infec-tion were investigated. Gene Set Variation Analysis (GSVA) was used to survey the mechanisms underlying OSCC carcinogenesis with and without C. albicans infection. The results revealed spe-cific cell clusters contributing to OSCC carcinogenesis with and without C. albicans infection. The major mechanisms involved in OSCC carcinogenesis without C. albicans infection are the IL2/STAT5, TNFα/NFκB, and TGFβ signaling pathways, whereas those involved in OSCC carcinogenesis with C. albicans infection are the KRAS signaling pathway and E2F target down-stream genes. Finally, stratifin (SFN) was validated to be a specific biomarker of OSCC with C. albicans infection. Thus, the detailed mechanism underlying OSCC carcinogenesis with C. albicans infection was determined and identified the treatment biomarker with potential precision medicine applications.
Keywords: Candida albicans; oral potentially malignant disorder; oral squamous cell carcinoma; single-cell RNA sequencing analysis; tumor heterogeneity; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
- MOHW107-TDU-B-212-114013, MOHW109-TDU-B-212-134016, MOHW110-TDU-B-212-144013, and MOHW111-TDU-B-221-114012/Ministry of Health and Welfare
- 109-2314-B-006-013-, 109-2740-B-400-002-, 108-2314-B-006-018-, 106-2314-B-006-016-, and 104-2314-B-006-062-/Ministry of Science and Technology
- CA-109-PP-18/National Health Research Institutes
- NCKUH-11104013, NCKUH-10902064, NCKUH-10604032, NCKUH-10406031/National Cheng Kung University Hospital
- 2021 UMP-NCKU/National Cheng Kung University
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

