Identification of ATP2B4 Regulatory Element Containing Functional Genetic Variants Associated with Severe Malaria
- PMID: 35563239
- PMCID: PMC9101746
- DOI: 10.3390/ijms23094849
Identification of ATP2B4 Regulatory Element Containing Functional Genetic Variants Associated with Severe Malaria
Abstract
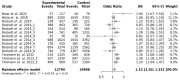
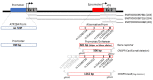
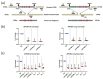
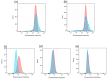
Genome-wide association studies for severe malaria (SM) have identified 30 genetic variants mostly located in non-coding regions. Here, we aimed to identify potential causal genetic variants located in these loci and demonstrate their functional activity. We systematically investigated the regulatory effect of the SNPs in linkage disequilibrium (LD) with the malaria-associated genetic variants. Annotating and prioritizing genetic variants led to the identification of a regulatory region containing five ATP2B4 SNPs in LD with rs10900585. We found significant associations between SM and rs10900585 and our candidate SNPs (rs11240734, rs1541252, rs1541253, rs1541254, and rs1541255) in a Senegalese population. Then, we demonstrated that both individual SNPs and the combination of SNPs had regulatory effects. Moreover, CRISPR/Cas9-mediated deletion of this region decreased ATP2B4 transcript and protein levels and increased Ca2+ intracellular concentration in the K562 cell line. Our data demonstrate that severe malaria-associated genetic variants alter the expression of ATP2B4 encoding a plasma membrane calcium-transporting ATPase 4 (PMCA4) expressed on red blood cells. Altering the activity of this regulatory element affects the risk of SM, likely through calcium concentration effect on parasitaemia.
Keywords: ATP2B4; CRISPR-cas9; SNP; calcium; enhancer; functional genomics; gene reporter; malaria; promoter; regulatory element.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this article.
Figures





References
MeSH terms
Substances
Grants and funding
- African Higher Education Centers of Excellence project/Cheikh Anta Diop University
- Grant to finance Alassane Thiam's stay at TAGC in Marseille/Institut Pasteur
- Samia Nisar's PhD fellowship/Higher Education Commission
- Florian Rosier's PhD fellowship/French ministry of research
- Funding for research/Inserm
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

