Photoperiod-Dependent Expression of MicroRNA in Drosophila
- PMID: 35563325
- PMCID: PMC9100521
- DOI: 10.3390/ijms23094935
Photoperiod-Dependent Expression of MicroRNA in Drosophila
Abstract
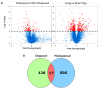
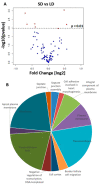
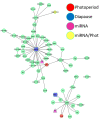
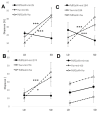
Like many other insects in temperate regions, Drosophila melanogaster exploits the photoperiod shortening that occurs during the autumn as an important cue to trigger a seasonal response. Flies survive the winter by entering a state of reproductive arrest (diapause), which drives the relocation of resources from reproduction to survival. Here, we profiled the expression of microRNA (miRNA) in long and short photoperiods and identified seven differentially expressed miRNAs (dme-mir-2b, dme-mir-11, dme-mir-34, dme-mir-274, dme-mir-184, dme-mir-184*, and dme-mir-285). Misexpression of dme-mir-2b, dme-mir-184, and dme-mir-274 in pigment-dispersing, factor-expressing neurons largely disrupted the normal photoperiodic response, suggesting that these miRNAs play functional roles in photoperiodic timing. We also analyzed the targets of photoperiodic miRNA by both computational predication and by Argonaute-1-mediated immunoprecipitation of long- and short-day RNA samples. Together with global transcriptome profiling, our results expand existing data on other Drosophila species, identifying genes and pathways that are differentially regulated in different photoperiods and reproductive status. Our data suggest that post-transcriptional regulation by miRNA is an important facet of photoperiodic timing.
Keywords: Drosophila; RNA immunoprecipitation; diapause; microRNA; photoperiodism; seasonal timing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Saunders D.S., Lewis R.D., Warman G.R. Photoperiodic induction of diapause: Opening the black box. Physiol. Entomol. 2004;29:1–15. doi: 10.1111/j.1365-3032.2004.0369.x. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

