HPV16 and HPV18 Genome Structure, Expression, and Post-Transcriptional Regulation
- PMID: 35563334
- PMCID: PMC9105396
- DOI: 10.3390/ijms23094943
HPV16 and HPV18 Genome Structure, Expression, and Post-Transcriptional Regulation
Erratum in
-
Correction: Yu et al. HPV16 and HPV18 Genome Structure, Expression, and Post-Transcriptional Regulation. Int. J. Mol. Sci. 2022, 23, 4943.Int J Mol Sci. 2022 Jul 18;23(14):7903. doi: 10.3390/ijms23147903. Int J Mol Sci. 2022. PMID: 35887403 Free PMC article.
Abstract
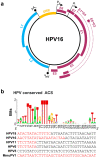
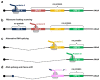
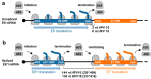
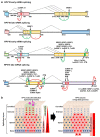
Human papillomaviruses (HPV) are a group of small non-enveloped DNA viruses whose infection causes benign tumors or cancers. HPV16 and HPV18, the two most common high-risk HPVs, are responsible for ~70% of all HPV-related cervical cancers and head and neck cancers. The expression of the HPV genome is highly dependent on cell differentiation and is strictly regulated at the transcriptional and post-transcriptional levels. Both HPV early and late transcripts differentially expressed in the infected cells are intron-containing bicistronic or polycistronic RNAs bearing more than one open reading frame (ORF), because of usage of alternative viral promoters and two alternative viral RNA polyadenylation signals. Papillomaviruses proficiently engage alternative RNA splicing to express individual ORFs from the bicistronic or polycistronic RNA transcripts. In this review, we discuss the genome structures and the updated transcription maps of HPV16 and HPV18, and the latest research advances in understanding RNA cis-elements, intron branch point sequences, and RNA-binding proteins in the regulation of viral RNA processing. Moreover, we briefly discuss the epigenetic modifications, including DNA methylation and possible APOBEC-mediated genome editing in HPV infections and carcinogenesis.
Keywords: RNA polyadenylation; RNA splicing; epigenetic modification; genome structure; papillomaviruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Pastrana D.V., Peretti A., Welch N.L., Borgogna C., Olivero C., Badolato R., Notarangelo L.D., Gariglio M., FitzGerald P.C., McIntosh C.E., et al. Metagenomic Discovery of 83 New Human Papillomavirus Types in Patients with Immunodeficiency. mSphere. 2018;3:e00645-18. doi: 10.1128/mSphereDirect.00645-18. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

