Transcriptomic Profiling of Peripheral Edge of Lesions to Elucidate the Pathogenesis of Psoriasis Vulgaris
- PMID: 35563374
- PMCID: PMC9101153
- DOI: 10.3390/ijms23094983
Transcriptomic Profiling of Peripheral Edge of Lesions to Elucidate the Pathogenesis of Psoriasis Vulgaris
Abstract
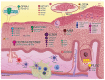
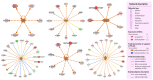
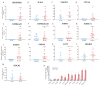
Elucidating transcriptome in the peripheral edge of the lesional (PE) skin could provide a better understanding of the molecules or signalings that intensify inflammation in the PE skin. Full-thickness biopsies of PE skin and uninvolved (UN) skin were obtained from psoriasis patients for RNA-seq. Several potential differentially expressed genes (DEGs) in the PE skin compared to those in the UN skin were identified. These DEGs enhanced functions such as angiogenesis, growth of epithelial tissue, chemotaxis and homing of cells, growth of connective tissues, and degranulation of myeloid cells beneath the PE skin. Moreover, the canonical pathways of IL-17A, IL-6, and IL-22 signaling were enriched by the DEGs. Finally, we proposed that inflammation in the PE skin might be driven by the IL-36/TLR9 axis or IL-6/Th17 axis and potentiated by IL-36α, IL-36γ, IL-17C, IL-8, S100A7, S100A8, S100A9, S100A15, SERPINB4, and hBD-2. Along with IL-36α, IL-17C, and IκBζ, ROCK2 could be an equally important factor in the pathogenesis of psoriasis, which may involve self-sustaining circuits between innate and adaptive immune responses via regulation of IL-36α and IL-36γ expression. Our finding provides new insight into signaling pathways in PE skin, which could lead to the discovery of new psoriasis targets.
Keywords: RNA sequencing; chronic skin diseases; cytokines; gene; inflammation.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

