Ubiquitin and Ubiquitin-like Proteins in Cancer, Neurodegenerative Disorders, and Heart Diseases
- PMID: 35563444
- PMCID: PMC9105348
- DOI: 10.3390/ijms23095053
Ubiquitin and Ubiquitin-like Proteins in Cancer, Neurodegenerative Disorders, and Heart Diseases
Abstract
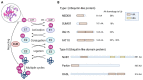
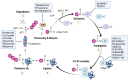
Post-translational modification (PTM) is an essential mechanism for enhancing the functional diversity of proteins and adjusting their signaling networks. The reversible conjugation of ubiquitin (Ub) and ubiquitin-like proteins (Ubls) to cellular proteins is among the most prevalent PTM, which modulates various cellular and physiological processes by altering the activity, stability, localization, trafficking, or interaction networks of its target molecules. The Ub/Ubl modification is tightly regulated as a multi-step enzymatic process by enzymes specific to this family. There is growing evidence that the dysregulation of Ub/Ubl modifications is associated with various diseases, providing new targets for drug development. In this review, we summarize the recent progress in understanding the roles and therapeutic targets of the Ub and Ubl systems in the onset and progression of human diseases, including cancer, neurodegenerative disorders, and heart diseases.
Keywords: disease association and progression; post-translational modification; ubiquitin; ubiquitin-like proteins; ubiquitin-proteasome system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

