Focused Design of Novel Cyclic Peptides Endowed with GABARAP-Inhibiting Activity
- PMID: 35563459
- PMCID: PMC9100651
- DOI: 10.3390/ijms23095070
Focused Design of Novel Cyclic Peptides Endowed with GABARAP-Inhibiting Activity
Abstract
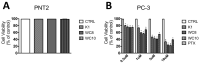
(1) Background: Disfunctions in autophagy machinery have been identified in various conditions, including neurodegenerative diseases, cancer, and inflammation. Among mammalian autophagy proteins, the Atg8 family member GABARAP has been shown to be greatly involved in the autophagy process of prostate cancer cells, supporting the idea that GABARAP inhibitors could be valuable tools to fight the progression of tumors. (2) Methods: In this paper, starting from the X-ray crystal structure of GABARAP in a complex with an AnkirinB-LIR domain, we identify two new peptides by applying in silico drug design techniques. The two ligands are synthesized, biophysically assayed, and biologically evaluated to ascertain their potential anticancer profile. (3) Results: Two cyclic peptides (WC8 and WC10) displayed promising biological activity, high conformational stability (due to the presence of disulfide bridges), and Kd values in the low micromolar range. The anticancer assays, performed on PC-3 cells, proved that both peptides exhibit antiproliferative effects comparable to those of peptide K1, a known GABARAP inhibitor. (4) Conclusions: WC8 and WC10 can be considered new GABARAP inhibitors to be employed as pharmacological tools or even templates for the rational design of new small molecules.
Keywords: Atg8; GABARAP inhibitors; LIR motif; PC-3; autophagy; cancer; peptide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Computational Design of Novel Cyclic Peptides Endowed with Autophagy-Inhibiting Activity on Cancer Cell Lines.Int J Mol Sci. 2024 Apr 24;25(9):4622. doi: 10.3390/ijms25094622. Int J Mol Sci. 2024. PMID: 38731842 Free PMC article.
-
The crystal structure of the FAM134B-GABARAP complex provides mechanistic insights into the selective binding of FAM134 to the GABARAP subfamily.FEBS Open Bio. 2022 Jan;12(1):320-331. doi: 10.1002/2211-5463.13340. Epub 2021 Dec 9. FEBS Open Bio. 2022. PMID: 34854256 Free PMC article.
-
An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5.Autophagy. 2020 Feb;16(2):256-270. doi: 10.1080/15548627.2019.1606637. Epub 2019 Apr 28. Autophagy. 2020. PMID: 30990354 Free PMC article.
-
Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors.J Mol Biol. 2020 Jan 3;432(1):80-103. doi: 10.1016/j.jmb.2019.07.016. Epub 2019 Jul 13. J Mol Biol. 2020. PMID: 31310766 Review.
-
Mechanisms of Selective Autophagy.Annu Rev Cell Dev Biol. 2021 Oct 6;37:143-169. doi: 10.1146/annurev-cellbio-120219-035530. Epub 2021 Jun 21. Annu Rev Cell Dev Biol. 2021. PMID: 34152791 Review.
Cited by
-
Computational Design of Novel Cyclic Peptides Endowed with Autophagy-Inhibiting Activity on Cancer Cell Lines.Int J Mol Sci. 2024 Apr 24;25(9):4622. doi: 10.3390/ijms25094622. Int J Mol Sci. 2024. PMID: 38731842 Free PMC article.
-
A new perspective on the autophagic and non-autophagic functions of the GABARAP protein family: a potential therapeutic target for human diseases.Mol Cell Biochem. 2024 Jun;479(6):1415-1441. doi: 10.1007/s11010-023-04800-5. Epub 2023 Jul 13. Mol Cell Biochem. 2024. PMID: 37440122 Review.
-
Progress of the "Molecular Informatics" Section in 2022.Int J Mol Sci. 2023 May 29;24(11):9442. doi: 10.3390/ijms24119442. Int J Mol Sci. 2023. PMID: 37298393 Free PMC article.
-
Linear motifs regulating protein secretion, sorting and autophagy in Leishmania parasites are diverged with respect to their host equivalents.PLoS Comput Biol. 2024 Feb 16;20(2):e1011902. doi: 10.1371/journal.pcbi.1011902. eCollection 2024 Feb. PLoS Comput Biol. 2024. PMID: 38363808 Free PMC article.
-
FYCO1 Peptide Analogs: Design and Characterization of Autophagy Inhibitors as Co-Adjuvants in Taxane Chemotherapy of Prostate Cancer.Int J Mol Sci. 2025 Jun 3;26(11):5365. doi: 10.3390/ijms26115365. Int J Mol Sci. 2025. PMID: 40508175 Free PMC article.
References
-
- Wright T.J., McKee C., Birch-Machin M.A., Ellis R., Armstrong J.L., Lovat P.E. Increasing the therapeutic efficacy of docetaxel for cutaneous squamous cell carcinoma through the combined inhibition of phosphatidylinositol 3-kinase/AKT signalling and autophagy. Clin. Exp. Dermatol. 2013;38:421–423. doi: 10.1111/ced.12138. - DOI - PubMed
-
- Rosenfeld M.R., Ye X., Supko J.G., Desideri S., Grossman S.A., Brem S., Mikkelson T., Wang D., Chang Y.C., Hu J., et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10:1359–1368. doi: 10.4161/auto.28984. - DOI - PMC - PubMed

