Fine-Tuning Modulation of Oxidation-Mediated Posttranslational Control of Bradyrhizobium diazoefficiens FixK2 Transcription Factor
- PMID: 35563511
- PMCID: PMC9104804
- DOI: 10.3390/ijms23095117
Fine-Tuning Modulation of Oxidation-Mediated Posttranslational Control of Bradyrhizobium diazoefficiens FixK2 Transcription Factor
Abstract
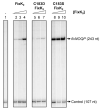
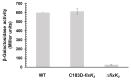
FixK2 is a CRP/FNR-type transcription factor that plays a central role in a sophisticated regulatory network for the anoxic, microoxic and symbiotic lifestyles of the soybean endosymbiont Bradyrhizobium diazoefficiens. Aside from the balanced expression of the fixK2 gene under microoxic conditions (induced by the two-component regulatory system FixLJ and negatively auto-repressed), FixK2 activity is posttranslationally controlled by proteolysis, and by the oxidation of a singular cysteine residue (C183) near its DNA-binding domain. To simulate the permanent oxidation of FixK2, we replaced C183 for aspartic acid. Purified C183D FixK2 protein showed both low DNA binding and in vitro transcriptional activation from the promoter of the fixNOQP operon, required for respiration under symbiosis. However, in a B. diazoefficiens strain coding for C183D FixK2, expression of a fixNOQP'-'lacZ fusion was similar to that in the wild type, when both strains were grown microoxically. The C183D FixK2 encoding strain also showed a wild-type phenotype in symbiosis with soybeans, and increased fixK2 gene expression levels and FixK2 protein abundance in cells. These two latter observations, together with the global transcriptional profile of the microoxically cultured C183D FixK2 encoding strain, suggest the existence of a finely tuned regulatory strategy to counterbalance the oxidation-mediated inactivation of FixK2 in vivo.
Keywords: CRP/FNR proteins; in vitro transcription; microarrays; microoxia; protein–DNA interaction; rhizobia; symbiosis.
Conflict of interest statement
The authors declare that no conflict of interest exist.
Figures





Similar articles
-
Dissection of FixK2 protein-DNA interaction unveils new insights into Bradyrhizobium diazoefficiens lifestyles control.Environ Microbiol. 2021 Oct;23(10):6194-6209. doi: 10.1111/1462-2920.15661. Epub 2021 Jul 28. Environ Microbiol. 2021. PMID: 34227211
-
Transcription activation in vitro by the Bradyrhizobium japonicum regulatory protein FixK2.J Bacteriol. 2005 May;187(10):3329-38. doi: 10.1128/JB.187.10.3329-3338.2005. J Bacteriol. 2005. PMID: 15866917 Free PMC article.
-
Regulation of Polyhydroxybutyrate Synthesis in the Soil Bacterium Bradyrhizobium diazoefficiens.Appl Environ Microbiol. 2016 Jun 30;82(14):4299-4308. doi: 10.1128/AEM.00757-16. Print 2016 Jul 15. Appl Environ Microbiol. 2016. PMID: 27208130 Free PMC article.
-
An Integrated Systems Approach Unveils New Aspects of Microoxia-Mediated Regulation in Bradyrhizobium diazoefficiens.Front Microbiol. 2019 May 7;10:924. doi: 10.3389/fmicb.2019.00924. eCollection 2019. Front Microbiol. 2019. PMID: 31134003 Free PMC article.
-
A multitude of CRP/FNR-like transcription proteins in Bradyrhizobium japonicum.Biochem Soc Trans. 2006 Feb;34(Pt 1):156-9. doi: 10.1042/BST0340156. Biochem Soc Trans. 2006. PMID: 16417509 Review.
Cited by
-
Independent Component Analysis Reveals the Transcriptional Regulatory Modules in Bradyrhizobium diazoefficiens USDA110.Int J Mol Sci. 2023 Aug 8;24(16):12544. doi: 10.3390/ijms241612544. Int J Mol Sci. 2023. PMID: 37628727 Free PMC article.
-
Pleiotropic Effects of PhaR Regulator in Bradyrhizobium diazoefficiens Microaerobic Metabolism.Int J Mol Sci. 2024 Feb 10;25(4):2157. doi: 10.3390/ijms25042157. Int J Mol Sci. 2024. PMID: 38396833 Free PMC article.
-
Molecular Advances in Microbial Metabolism.Int J Mol Sci. 2023 Apr 28;24(9):8015. doi: 10.3390/ijms24098015. Int J Mol Sci. 2023. PMID: 37175720 Free PMC article.
-
Surface Plasmon Resonance as a Tool to Elucidate the Molecular Determinants of Key Transcriptional Regulators Controlling Rhizobial Lifestyles.Methods Mol Biol. 2024;2751:145-163. doi: 10.1007/978-1-0716-3617-6_10. Methods Mol Biol. 2024. PMID: 38265715
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

