Tissue Transglutaminase Knock-Out Preadipocytes and Beige Cells of Epididymal Fat Origin Possess Decreased Mitochondrial Functions Required for Thermogenesis
- PMID: 35563567
- PMCID: PMC9105016
- DOI: 10.3390/ijms23095175
Tissue Transglutaminase Knock-Out Preadipocytes and Beige Cells of Epididymal Fat Origin Possess Decreased Mitochondrial Functions Required for Thermogenesis
Abstract
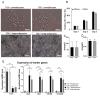
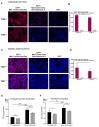
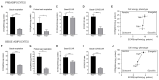
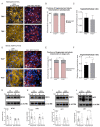
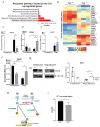
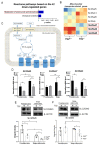
Beige adipocytes with thermogenic function are activated during cold exposure in white adipose tissue through the process of browning. These cells, similar to brown adipocytes, dissipate stored chemical energy in the form of heat with the help of uncoupling protein 1 (UCP1). Recently, we have shown that tissue transglutaminase (TG2) knock-out mice have decreased cold tolerance in parallel with lower utilization of their epididymal adipose tissue and reduced browning. To learn more about the thermogenic function of this fat depot, we isolated preadipocytes from the epididymal adipose tissue of wild-type and TG2 knock-out mice and differentiated them in the beige direction. Although differentiation of TG2 knock-out preadipocytes is phenotypically similar to the wild-type cells, the mitochondria of the knock-out beige cells have multiple impairments including an altered electron transport system generating lower electrochemical potential difference, reduced oxygen consumption, lower UCP1 protein content, and a higher portion of fragmented mitochondria. Most of these differences are present in preadipocytes as well, and the differentiation process cannot overcome the functional disadvantages completely. TG2 knock-out beige adipocytes produce more iodothyronine deiodinase 3 (DIO3) which may inactivate thyroid hormones required for the establishment of optimal mitochondrial function. The TG2 knock-out preadipocytes and beige cells are both hypometabolic as compared with the wild-type controls which may also be explained by the lower expression of solute carrier proteins SLC25A45, SLC25A47, and SLC25A42 which transport acylcarnitine, Co-A, and amino acids into the mitochondrial matrix. As a consequence, the mitochondria in TG2 knock-out beige adipocytes probably cannot reach the energy-producing threshold required for normal thermogenic functions, which may contribute to the decreased cold tolerance of TG2 knock-out mice.
Keywords: DIO3; SLC25A45; beige adipocytes; browning; uncoupling protein-1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Folk J.E., Finlayson J.S. The epsilon-(gamma-glutamyl) lysine crosslink and the catalytic role of transglutaminases. Adv. Protein Chem. 1977;31:1–133. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

