Mechanisms of Resistance to Spot Blotch in Yunnan Iron Shell Wheat Based on Metabolome and Transcriptomics
- PMID: 35563578
- PMCID: PMC9104156
- DOI: 10.3390/ijms23095184
Mechanisms of Resistance to Spot Blotch in Yunnan Iron Shell Wheat Based on Metabolome and Transcriptomics
Abstract
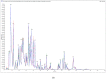
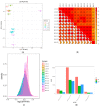
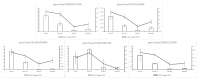
Spot blotch (SB) is a fungal disease that threatens wheat yield and quality. Presently, the molecular mechanism against SB is unclear. In this study, the resistant variety Zhenkang iron shell wheat (Yunmai 0030) and susceptible variety Lincang iron shell wheat (Yunmai 0608) were selected by identifying SB of Yunnan iron shell wheat. The metabolome and transcriptome of leaves of two varieties at different positions were detected using the systemic acquired resistance theory to investigate the molecular and physiological changes in Yunnan iron shell wheat under SB stress. We found that the genes and metabolites related to benzoxazinoid biosynthesis and arginine and proline metabolism were highly enriched after infection with leaf blight. The enriched differential metabolites mainly included phenolic acids, alkaloids, and flavonoids. We further observed that DIBOA- and DIMBOA-glucoside positively affected iron shell wheat resistance to leaf blight and proline and its derivatives were important for plant self-defense. Furthermore, we confirmed that the related metabolites in benzoxazinoid biosynthesis and arginine and proline metabolism positively affected Triticum aestivum ssp. resistance to SB. This study provides new insights into the dynamic physiological changes of wheat in response to SB, helps us better understand the mechanism of resistance to SB, and contributes to the breeding and utilization of resistant varieties.
Keywords: Triticum aestivum ssp.; benzoxazinoid biosynthesis; metabolome; spot blotch; transcriptome.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures











References
-
- Wang X., Dong L., Hu J., Pang Y., Hu L., Xiao G., Ma X., Kong X., Jia J., Wang H., et al. Dissecting genetic loci affecting grain morphological traits to improve grain weight via nested association mapping. Theor. Appl. Genet. 2019;132:3115–3128. doi: 10.1007/s00122-019-03410-4. - DOI - PMC - PubMed
-
- Li J., Jiang Y., Yao F., Long L., Wang Y., Wu Y., Li H., Wang J., Jiang Q., Kang H., et al. Genome-Wide Association Study Reveals the Genetic Architecture of Stripe Rust Resistance at the Adult Plant Stage in Chinese Endemic Wheat. Front. Plant Sci. 2020;11:625. doi: 10.3389/fpls.2020.00625. - DOI - PMC - PubMed
-
- Dong Y., Zheng D., Qiao D., Zeng X., En Z., Chen X. Expedition and investigation of Yunnan wheat (Triticum aestivum ssp. yunnanense King) Acta Agronom. Sin. 1981;7:145–152. (In Chinese)
-
- Dong Y.S., Zheng D.S. Chinese Wheat Genetic Resources. China Agriculture Press; Beijing, China: 1999. pp. 53–56.
-
- Zhang X.Y., Pang B.S., You G.X., Wang L.F., Jia J.Z., Dong Y.C. Allelic Variation and Genetic Diversity at Glu-1 Loci in Chinese Wheat (Triticum aestivum L.) Germplasms. Agric. Sci. China. 2002;1:1074–1082.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

