Polysialylation in a DISC1 Mutant Mouse
- PMID: 35563598
- PMCID: PMC9102787
- DOI: 10.3390/ijms23095207
Polysialylation in a DISC1 Mutant Mouse
Abstract
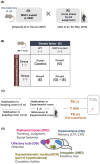
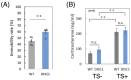
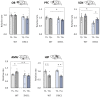
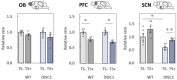
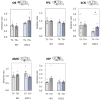
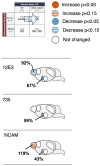
Schizophrenia is a serious psychiatric disorder that affects the social life of patients. Psychiatric disorders are caused by a complex combination of genetic (G) and environmental (E) factors. Polysialylation represents a unique posttranslational modification of a protein, and such changes in neural cell adhesion molecules (NCAMs) have been reported in postmortem brains from patients with psychiatric disorders. To understand the G × E effect on polysialylated NCAM expression, in this study, we performed precise measurements of polySia and NCAM using a disrupted-in-schizophrenia 1 (DISC1)-mutant mouse (G), a mouse model of schizophrenia, under acute stress conditions (E). This is the first study to reveal a lower number and smaller length of polySia in the suprachiasmatic nucleus of DISC1 mutants relative to those in wild-type (WT) mice. In addition, an analysis of polySia and NCAM responses to acute stress in five brain regions (olfactory bulb, prefrontal cortex, suprachiasmatic nucleus, amygdala, and hippocampus) revealed that the pattern of changes in these responses in WT mice and DISC1 mutants differed by region. These differences could indicate the vulnerability of DISC1 mutants to stress.
Keywords: DISC1; NCAM; acute stress; environmental factor; genetic factor; mental disorder; polysialic acid; polysialyltransferase; schizophrenia; tail suspension test.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

