Genome-Wide Identification of the Eucalyptus urophylla GATA Gene Family and Its Diverse Roles in Chlorophyll Biosynthesis
- PMID: 35563644
- PMCID: PMC9102942
- DOI: 10.3390/ijms23095251
Genome-Wide Identification of the Eucalyptus urophylla GATA Gene Family and Its Diverse Roles in Chlorophyll Biosynthesis
Abstract
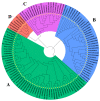
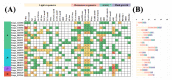
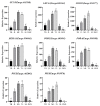
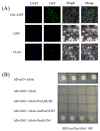
GATA transcription factors have been demonstrated to play key regulatory roles in plant growth, development, and hormonal response. However, the knowledge concerning the evolution of GATA genes in Eucalyptus urophylla and their trans-regulatory interaction is indistinct. Phylogenetic analysis and study of conserved motifs, exon structures, and expression patterns resolved the evolutionary relationships of these GATA proteins. Phylogenetic analysis showed that EgrGATAs are broadly distributed in four subfamilies. Cis-element analysis of promoters revealed that EgrGATA genes respond to light and are influenced by multiple hormones and abiotic stresses. Transcriptome analysis revealed distinct temporal and spatial expression patterns of EgrGATA genes in various tissues of E. urophylla S.T.Blake, which was confirmed by real-time quantitative PCR (RT-qPCR). Further research revealed that EurGNC and EurCGA1 were localized in the nucleus, and EurGNC directly binds to the cis-element of the EurGUN5 promoter, implying its potential roles in the regulation of chlorophyll synthesis. This comprehensive study provides new insights into the evolution of GATAs and could help to improve the photosynthetic assimilation and vegetative growth of E. urophylla at the genetic level.
Keywords: Eucalyptus; GATA; evolution; photosynthesis; transcription factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

