Generation of Isogenic hiPSCs with Targeted Edits at Multiple Intronic SNPs to Study the Effects of the Type 2 Diabetes Associated KCNQ1 Locus in American Indians
- PMID: 35563754
- PMCID: PMC9102014
- DOI: 10.3390/cells11091446
Generation of Isogenic hiPSCs with Targeted Edits at Multiple Intronic SNPs to Study the Effects of the Type 2 Diabetes Associated KCNQ1 Locus in American Indians
Abstract
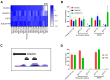
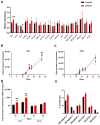
The top genetic association signal for type 2 diabetes (T2D) in Southwestern American Indians maps to intron 15 of KCNQ1, an imprinted gene. We aim to understand the biology whereby variation at this locus affects T2D specifically in this genomic background. To do so, we obtained human induced pluripotent stem cells (hiPSC) derived from American Indians. Using these iPSCs, we show that imprinting of KCNQ1 and CDKN1C during pancreatic islet-like cell generation from iPSCs is consistent with known imprinting patterns in fetal pancreas and adult islets and therefore is an ideal model system to study this locus. In this report, we detail the use of allele-specific guide RNAs and CRISPR to generate isogenic hiPSCs that differ only at multiple T2D associated intronic SNPs at this locus which can be used to elucidate their functional effects. Characterization of these isogenic hiPSCs identified a few aberrant cell lines; namely cell lines with large hemizygous deletions in the putative functional region of KCNQ1 and cell lines hypomethylated at the KCNQ1OT1 promoter. Comparison of an isogenic cell line with a hemizygous deletion to the parental cell line identified CDKN1C and H19 as differentially expressed during the endocrine progenitor stage of pancreatic-islet development.
Keywords: CDKN1C; H19; KCNQ1; allele-specific guide RNA; hiPSCs; pancreatic-beta like cells; type 2 diabetes.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. - DOI - PMC - PubMed
-
- Adeyemo A.A., Zaghloul N.A., Chen G., Doumatey A.P., Leitch C.C., Hostelley T.L., Nesmith J.E., Zhou J., Bentley A.R., Shriner D., et al. ZRANB3 is an African-specific type 2 diabetes locus associated with beta-cell mass and insulin response. Nat. Commun. 2019;10:3195. doi: 10.1038/s41467-019-10967-7. - DOI - PMC - PubMed
-
- Hanson R.L., Muller Y.L., Kobes S., Guo T., Bian L., Ossowski V., Wiedrich K., Sutherland J., Wiedrich C., Mahkee D., et al. A Genome-Wide Association Study in American Indians Implicates DNER as a Susceptibility Locus for Type 2 Diabetes. Diabetes. 2014;63:369–376. doi: 10.2337/db13-0416. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

