XRN2 Is Required for Cell Motility and Invasion in Glioblastomas
- PMID: 35563787
- PMCID: PMC9100175
- DOI: 10.3390/cells11091481
XRN2 Is Required for Cell Motility and Invasion in Glioblastomas
Abstract
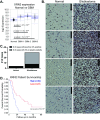
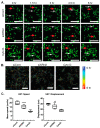
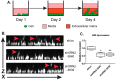
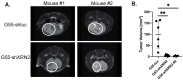
One of the major obstacles in treating brain cancers, particularly glioblastoma multiforme, is the occurrence of secondary tumor lesions that arise in areas of the brain and are inoperable while obtaining resistance to current therapeutic agents. Thus, gaining a better understanding of the cellular factors that regulate glioblastoma multiforme cellular movement is imperative. In our study, we demonstrate that the 5'-3' exoribonuclease XRN2 is important to the invasive nature of glioblastoma. A loss of XRN2 decreases cellular speed, displacement, and movement through a matrix of established glioblastoma multiforme cell lines. Additionally, a loss of XRN2 abolishes tumor formation in orthotopic mouse xenograft implanted with G55 glioblastoma multiforme cells. One reason for these observations is that loss of XRN2 disrupts the expression profile of several cellular factors that are important for tumor invasion in glioblastoma multiforme cells. Importantly, XRN2 mRNA and protein levels are elevated in glioblastoma multiforme patient samples. Elevation in XRN2 mRNA also correlates with poor overall patient survival. These data demonstrate that XRN2 is an important cellular factor regulating one of the major obstacles in treating glioblastomas and is a potential molecular target that can greatly enhance patient survival.
Keywords: XRN2; cell motility; invasion; tumor progression.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- Minniti G., Muni R., Lanzetta G., Marchetti P., Enrici R.M. Chemotherapy for Glioblastoma: Current Treatment and Future Perspectives for Cytotoxic and Targeted Agents. Anticancer. Res. 2009;29:5171–5184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

