The Cytoskeleton Effectors Rho-Kinase (ROCK) and Mammalian Diaphanous-Related (mDia) Formin Have Dynamic Roles in Tumor Microtube Formation in Invasive Glioblastoma Cells
- PMID: 35563863
- PMCID: PMC9103681
- DOI: 10.3390/cells11091559
The Cytoskeleton Effectors Rho-Kinase (ROCK) and Mammalian Diaphanous-Related (mDia) Formin Have Dynamic Roles in Tumor Microtube Formation in Invasive Glioblastoma Cells
Abstract
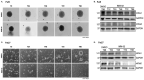
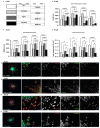
Glioblastoma (GBM) is a progressive and lethal brain cancer. Malignant control of actin and microtubule cytoskeletal mechanics facilitates two major GBM therapeutic resistance strategies-diffuse invasion and tumor microtube network formation. Actin and microtubule reorganization is controlled by Rho-GTPases, which exert their effects through downstream effector protein activation, including Rho-associated kinases (ROCK) 1 and 2 and mammalian diaphanous-related (mDia) formins (mDia1, 2, and 3). Precise spatial and temporal balancing of the activity between these effectors dictates cell shape, adhesion turnover, and motility. Using small molecules targeting mDia, we demonstrated that global agonism (IMM02) was superior to antagonism (SMIFH2) as anti-invasion strategies in GBM spheroids. Here, we use IDH-wild-type GBM patient-derived cell models and a novel semi-adherent in vitro system to investigate the relationship between ROCK and mDia in invasion and tumor microtube networks. IMM02-mediated mDia agonism disrupts invasion in GBM patient-derived spheroid models, in part by inducing mDia expression loss and tumor microtube network collapse. Pharmacological disruption of ROCK prevented invasive cell-body movement away from GBM spheres, yet induced ultralong, phenotypically abnormal tumor microtube formation. Simultaneously targeting mDia and ROCK did not enhance the anti-invasive/-tumor microtube effects of IMM02. Our data reveal that targeting mDia is a viable GBM anti-invasion/-tumor microtube networking strategy, while ROCK inhibition is contraindicated.
Keywords: Rho-kinase; actin; cytoskeleton; glioblastoma; invasion; mDia formin; tumor microtube.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L., et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

