Long-Distance Repression by Human Silencers: Chromatin Interactions and Phase Separation in Silencers
- PMID: 35563864
- PMCID: PMC9101175
- DOI: 10.3390/cells11091560
Long-Distance Repression by Human Silencers: Chromatin Interactions and Phase Separation in Silencers
Abstract
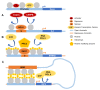
Three-dimensional genome organization represents an additional layer in the epigenetic regulation of gene expression. Active transcription controlled by enhancers or super-enhancers has been extensively studied. Enhancers or super-enhancers can recruit activators or co-activators to activate target gene expression through long-range chromatin interactions. Chromatin interactions and phase separation play important roles in terms of enhancer or super-enhancer functioning. Silencers are another major type of cis-regulatory element that can mediate gene regulation by turning off or reducing gene expression. However, compared to active transcription, silencer studies are still in their infancy. This review covers the current knowledge of human silencers, especially the roles of chromatin interactions and phase separation in silencers. This review also proposes future directions for human silencer studies.
Keywords: chromatin interactions; phase separation; silencers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Goode D.K., Obier N., Vijayabaskar M.S., Lie-A-Ling M., Lilly A.J., Hannah R., Lichtinger M., Batta K., Florkowska M., Patel R., et al. Dynamic Gene Regulatory Networks Drive Hematopoietic Specification and Differentiation. Dev. Cell. 2016;36:572–587. doi: 10.1016/j.devcel.2016.01.024. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

