Molecular Epidemiology of Antibiotic Resistance Genes and Virulence Factors in Multidrug-Resistant Escherichia coli Isolated from Rodents, Humans, Chicken, and Household Soils in Karatu, Northern Tanzania
- PMID: 35564782
- PMCID: PMC9102629
- DOI: 10.3390/ijerph19095388
Molecular Epidemiology of Antibiotic Resistance Genes and Virulence Factors in Multidrug-Resistant Escherichia coli Isolated from Rodents, Humans, Chicken, and Household Soils in Karatu, Northern Tanzania
Abstract
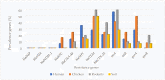
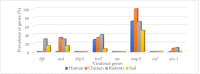
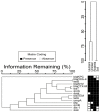
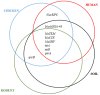
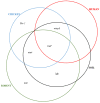
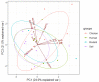
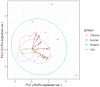
The interaction of rodents with humans and chicken in the household environment can facilitate transmission of multidrug-resistant (MDR) Escherichia coli (E. coli), causing infections that are difficult to treat. We investigated the presence of genes encoded for carbapenem, extended spectrum beta-lactamases (ESBL), tetracycline and quinolones resistance, and virulence among 50 MDR E. coli isolated from human (n = 14), chicken (n = 12), rodent (n = 10), and soil (n = 14) samples using multiplex polymerase chain reaction (PCR). Overall, the antimicrobial resistance genes (ARGs) detected were: blaTEM 23/50 (46%), blaCTX-M 13/50 (26%), tetA 23/50 (46%), tetB 7/50 (14%), qnrA 12/50 (24%), qnrB 4/50 (8%), blaOXA-48 6/50 (12%), and blaKPC 3/50 (6%), while blaIMP, blaVIM, and blaNDM-1 were not found. The virulence genes (VGs) found were: ompA 36/50 (72%), traT 13/50 (26%), east 9/50 (18%), bfp 5/50 (10%), eae 1/50 (2%), and stx-1 2/50 (4%), while hlyA and cnf genes were not detected. Resistance (blaTEM, blaCTX-M, blaSHV, tetA, tetB, and qnrA) and virulence (traT) genes were found in all sample sources while stx-1 and eae were only found in chicken and rodent isolates, respectively. Tetracycline resistance phenotypes correlated with genotypes tetA (r = 0.94), tetB (r = 0.90), blaKPC (r = 0.90; blaOXA-48 (r = 0.89), and qnrA (r = 0.96). ESBL resistance was correlated with genotypes blaKPC (r = 0.93), blaOXA-48 (r = 0.90), and qnrA (r = 0.96) resistance. Positive correlations were observed between resistance and virulence genes: qnrB and bfp (r = 0.63) also blaTEM, and traT (r = 0.51). Principal component analysis (PCA) indicated that tetA, tetB, blaTEM, blaCTX-M, qnrA, and qnrB genes contributed to tetracycline, cefotaxime, and quinolone resistance, respectively. While traT stx-1, bfp, ompA, east, and eae genes contributed to virulence of MDR E. coli isolates. The PCA ellipses show that isolates from rodents had more ARGs and virulence genes compared to those isolated from chicken, soil, and humans.
Keywords: E. coli; PCR; chicken; genes; humans; multidrug-resistant; rodents; soil.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Occurrence of virulence genes in multidrug-resistant Escherichia coli isolates from humans, animals, and the environment: One health perspective.PLoS One. 2025 Jan 24;20(1):e0317874. doi: 10.1371/journal.pone.0317874. eCollection 2025. PLoS One. 2025. PMID: 39854442 Free PMC article.
-
Multiple sequence types, virulence determinants and antimicrobial resistance genes in multidrug- and colistin-resistant Escherichia coli from agricultural and non-agricultural soils.Environ Pollut. 2021 Nov 1;288:117804. doi: 10.1016/j.envpol.2021.117804. Epub 2021 Jul 17. Environ Pollut. 2021. PMID: 34329068
-
High Levels of Resistance to Cephalosporins Associated with the Presence of Extended-Spectrum and AmpC β-Lactamases in Escherichia coli from Broilers in Southern Brazil.Microb Drug Resist. 2020 May;26(5):531-535. doi: 10.1089/mdr.2019.0050. Epub 2019 Nov 12. Microb Drug Resist. 2020. PMID: 31718408
-
Escherichia coli: An arduous voyage from commensal to Antibiotic-resistance.Microb Pathog. 2025 Jan;198:107173. doi: 10.1016/j.micpath.2024.107173. Epub 2024 Nov 27. Microb Pathog. 2025. PMID: 39608506 Review.
-
Antimicrobial resistance genes of Escherichia coli, a bacterium of "One Health" importance in South Africa: Systematic review and meta-analysis.AIMS Microbiol. 2023 Feb 13;9(1):75-89. doi: 10.3934/microbiol.2023005. eCollection 2023. AIMS Microbiol. 2023. PMID: 36891533 Free PMC article. Review.
Cited by
-
Unveiling Rare Pathogens and Antibiotic Resistance in Tanzanian Cholera Outbreak Waters.Microorganisms. 2023 Oct 4;11(10):2490. doi: 10.3390/microorganisms11102490. Microorganisms. 2023. PMID: 37894148 Free PMC article.
-
Bacteriophages as a Biocontrol Strategy to Prevent the Contamination of Meat Products with Escherichia coli - a Meta-Analysis.Pol J Microbiol. 2025 Jun 18;74(2):165-176. doi: 10.33073/pjm-2025-014. eCollection 2025 Jun 1. Pol J Microbiol. 2025. PMID: 40544515 Free PMC article.
-
Comprehensive genome catalog analysis of the resistome, virulome and mobilome in the wild rodent gut microbiota.NPJ Biofilms Microbiomes. 2025 Jun 11;11(1):101. doi: 10.1038/s41522-025-00746-2. NPJ Biofilms Microbiomes. 2025. PMID: 40500303 Free PMC article.
-
Synanthropic rodents and shrews are reservoirs of zoonotic bacterial pathogens and act as sentinels for antimicrobial resistance spillover in the environment: A study from Puducherry, India.One Health. 2024 May 17;18:100759. doi: 10.1016/j.onehlt.2024.100759. eCollection 2024 Jun. One Health. 2024. PMID: 38784598 Free PMC article.
-
Escherichia coli isolates from vegetable farms in Addis Ababa, Ethiopia: Antimicrobial susceptibility profile and associated resistance genetic markers.Food Sci Nutr. 2024 Mar 10;12(6):4122-4132. doi: 10.1002/fsn3.4071. eCollection 2024 Jun. Food Sci Nutr. 2024. PMID: 38873492 Free PMC article.
References
-
- Song Y., Yu L., Zhang Y., Dai Y., Wang P., Feng C., Liu M., Sun S., Xie Z., Wang F. Prevalence and characteristics of multidrug-resistant mcr-1-positive Escherichia coli isolates from broiler chickens in Tai’an, China. Poult. Sci. 2020;99:1117–1123. doi: 10.1016/j.psj.2019.10.044. - DOI - PMC - PubMed
-
- Shrivastava S.R., Shrivastava P.S., Ramasamy J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018;32:76–77. doi: 10.4103/jms.jms_25_17. - DOI
-
- Sarowska J., Futoma-Koloch B., Jama-Kmiecik A., Frej-Madrzak M., Ksiazczyk M., Bugla-Ploskonska G., Choroszy-Krol I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019;11:10. doi: 10.1186/s13099-019-0290-0. - DOI - PMC - PubMed
-
- Abd El-Baky R.M., Ibrahim R.A., Mohamed D.S., Ahmed E.F., Hashem Z.S. Prevalence of virulence genes and their association with antimicrobial resistance among pathogenic E. coli isolated from Egyptian patients with different clinical infections. Infect. Drug. Resist. 2020;13:1221–1236. doi: 10.2147/IDR.S241073. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

