Convolutional Neural Networks to Detect Vestibular Schwannomas on Single MRI Slices: A Feasibility Study
- PMID: 35565199
- PMCID: PMC9104481
- DOI: 10.3390/cancers14092069
Convolutional Neural Networks to Detect Vestibular Schwannomas on Single MRI Slices: A Feasibility Study
Abstract
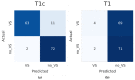
In this study. we aimed to detect vestibular schwannomas (VSs) in individual magnetic resonance imaging (MRI) slices by using a 2D-CNN. A pretrained CNN (ResNet-34) was retrained and internally validated using contrast-enhanced T1-weighted (T1c) MRI slices from one institution. In a second step, the model was externally validated using T1c- and T1-weighted (T1) slices from a different institution. As a substitute, bisected slices were used with and without tumors originating from whole transversal slices that contained part of the unilateral VS. The model predictions were assessed based on the categorical accuracy and confusion matrices. A total of 539, 94, and 74 patients were included for training, internal validation, and external T1c validation, respectively. This resulted in an accuracy of 0.949 (95% CI 0.935-0.963) for the internal validation and 0.912 (95% CI 0.866-0.958) for the external T1c validation. We suggest that 2D-CNNs might be a promising alternative to 2.5-/3D-CNNs for certain tasks thanks to the decreased demand for computational power and the fact that there is no need for segmentations. However, further research is needed on the difference between 2D-CNNs and more complex architectures.
Keywords: artificial intelligence; deep learning; machine learning; neuro-oncology; schwannoma; vestibular.
Conflict of interest statement
P.W. has a patent application titled ‘Method for detection of neurological abnormalities’. C.F. and F.E. received speaker honoraria from Accuray outside of the submitted work. The remaining authors declare no conflict of interest.
Figures






References
-
- Angra S., Ahuja S. Machine Learning and Its Applications: A Review; Proceedings of the 2017 International Conference on Big Data Analytics and Computational Intelligence (ICBDAC); Chirala, India. 23–25 March 2017; pp. 57–60.
-
- Tiwari A., Srivastava S., Pant M. Brain Tumor Segmentation and Classification from Magnetic Resonance Images: Review of Selected Methods from 2014 to 2019. Pattern Recognit. Lett. 2020;131:244–260. doi: 10.1016/j.patrec.2019.11.020. - DOI
-
- Ehret F., Kaul D., Clusmann H., Delev D., Kernbach J.M. Machine Learning-Based Radiomics in Neuro-Oncology. Acta Neurochir. Suppl. 2022;134:139–151. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

