Template-Independent Poly(A)-Tail Decay and RNASEL as Potential Cellular Biomarkers for Prostate Cancer Development
- PMID: 35565367
- PMCID: PMC9100668
- DOI: 10.3390/cancers14092239
Template-Independent Poly(A)-Tail Decay and RNASEL as Potential Cellular Biomarkers for Prostate Cancer Development
Abstract
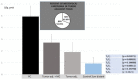
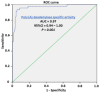
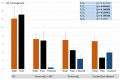
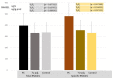
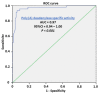
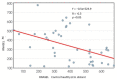
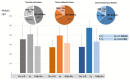
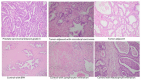
The post-transcriptional messenger RNA (mRNA) decay and turnover rate of the template-independent poly(A) tail, localized at the 3'-untranslated region (3'UTR) of mRNA, have been documented among subtle mechanisms of uncontrolled cancer tissue growth. The activity of Poly(A) deadenylase and the expression pattern of RNASEL have been examined. A total of 138 prostate tissue specimens from 46 PC patients (cancer specimens, corresponding adjacent surgically healthy tissues, and in their normal counterparts, at least 2 cm from carcinoma) were used. For the stratification prediction of healthy tissue transition into malignant phenotype, the enzyme activity of tumor-adjacent tissue was considered in relation to the presence of microfocal carcinoma. More than a four-times increase in specific enzyme activity (U/L g.prot) was registered in PC on account of both the dissociation of its inhibitor and genome reprogramming. The obtained ROC curve and Youden index showed that Poly(A) deadenylase identified PC with a sensitivity of 93.5% and a specificity of 94.6%. The RNASEL expression profile was raised significantly in PC, but the sensitivity was 40.5% and specificity was 86.9%. A significantly negative correlation between PC and control tissue counterparts with a higher expression pattern in lymphocyte-infiltrated samples were reported. In conclusion, significantly upregulated Poly(A) deadenylase activity may be a checkpoint for the transition of precancerous lesion to malignancy, while RNASEL may predict chronic inflammation.
Keywords: RNASEL; poly(A) deadenylase; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Serrano N.A., Anscher M.S. Favorable vs. Unfavorable Intermediate-Risk Prostate Cancer: A Review of the New Classification System and Its Impact on Treatment Recommendations. Oncology. 2016;30:229–236. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

