The Genetic and Molecular Analyses of RAD51C and RAD51D Identifies Rare Variants Implicated in Hereditary Ovarian Cancer from a Genetically Unique Population
- PMID: 35565380
- PMCID: PMC9104874
- DOI: 10.3390/cancers14092251
The Genetic and Molecular Analyses of RAD51C and RAD51D Identifies Rare Variants Implicated in Hereditary Ovarian Cancer from a Genetically Unique Population
Abstract
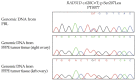
To identify candidate variants in RAD51C and RAD51D ovarian cancer (OC) predisposing genes by investigating French Canadians (FC) exhibiting unique genetic architecture. Candidates were identified by whole exome sequencing analysis of 17 OC families and 53 early-onset OC cases. Carrier frequencies were determined by the genetic analysis of 100 OC or HBOC families, 438 sporadic OC cases and 1025 controls. Variants of unknown function were assayed for their biological impact and/or cellular sensitivity to olaparib. RAD51C c.414G>C;p.Leu138Phe and c.705G>T;p.Lys235Asn and RAD51D c.137C>G;p.Ser46Cys, c.620C>T;p.Ser207Leu and c.694C>T;p.Arg232Ter were identified in 17.6% of families and 11.3% of early-onset cases. The highest carrier frequency was observed in OC families (1/44, 2.3%) and sporadic cases (15/438, 3.4%) harbouring RAD51D c.620C>T versus controls (1/1025, 0.1%). Carriers of c.620C>T (n = 7), c.705G>T (n = 2) and c.137C>G (n = 1) were identified in another 538 FC OC cases. RAD51C c.705G>T affected splicing by skipping exon four, while RAD51D p.Ser46Cys affected protein stability and conferred olaparib sensitivity. Genetic and functional assays implicate RAD51C c.705G>T and RAD51D c.137C>G as likely pathogenic variants in OC. The high carrier frequency of RAD51D c.620C>T in FC OC cases validates previous findings. Our findings further support the role of RAD51C and RAD51D in hereditary OC.
Keywords: French Canadian; RAD51C; RAD51D; genetic drift; ovarian cancer predisposing genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Mutation Analysis of the RAD51C and RAD51D Genes in High-Risk Ovarian Cancer Patients and Families from the Czech Republic.PLoS One. 2015 Jun 9;10(6):e0127711. doi: 10.1371/journal.pone.0127711. eCollection 2015. PLoS One. 2015. PMID: 26057125 Free PMC article.
-
Mutational analysis of RAD51C and RAD51D genes in hereditary breast and ovarian cancer families from Murcia (southeastern Spain).Eur J Med Genet. 2018 Jun;61(6):355-361. doi: 10.1016/j.ejmg.2018.01.015. Epub 2018 Feb 2. Eur J Med Genet. 2018. PMID: 29409816
-
Clinicopathological Characteristics of Ovarian and Breast Cancer in PALB2, RAD51C, and RAD51D Germline Pathogenic Variant Carriers.Genes (Basel). 2025 May 2;16(5):556. doi: 10.3390/genes16050556. Genes (Basel). 2025. PMID: 40428378 Free PMC article.
-
The Genetic Analyses of French Canadians of Quebec Facilitate the Characterization of New Cancer Predisposing Genes Implicated in Hereditary Breast and/or Ovarian Cancer Syndrome Families.Cancers (Basel). 2021 Jul 7;13(14):3406. doi: 10.3390/cancers13143406. Cancers (Basel). 2021. PMID: 34298626 Free PMC article. Review.
-
Germline mutations in RAD51C and RAD51D and hereditary predisposition to ovarian cancer.Klin Onkol. 2021 Winter;34(1):26-32. doi: 10.48095/ccko202126. Klin Onkol. 2021. PMID: 33657816 Review. English.
Cited by
-
Minigene Splicing Assays Identify 20 Spliceogenic Variants of the Breast/Ovarian Cancer Susceptibility Gene RAD51C.Cancers (Basel). 2022 Jun 15;14(12):2960. doi: 10.3390/cancers14122960. Cancers (Basel). 2022. PMID: 35740625 Free PMC article.
-
Molecular Genetic Characteristics of FANCI, a Proposed New Ovarian Cancer Predisposing Gene.Genes (Basel). 2023 Jan 20;14(2):277. doi: 10.3390/genes14020277. Genes (Basel). 2023. PMID: 36833203 Free PMC article.
-
Genetic analyses of DNA repair pathway associated genes implicate new candidate cancer predisposing genes in ancestrally defined ovarian cancer cases.Front Oncol. 2023 Mar 8;13:1111191. doi: 10.3389/fonc.2023.1111191. eCollection 2023. Front Oncol. 2023. PMID: 36969007 Free PMC article.
References
-
- Meindl A., Hellebrand H., Wiek C., Erven V., Wappenschmidt B., Niederacher D., Freund M., Lichtner P., Hartmann L., Schaal H., et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 2010;42:410–414. doi: 10.1038/ng.569. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

