Muscarinic Receptors Associated with Cancer
- PMID: 35565451
- PMCID: PMC9100020
- DOI: 10.3390/cancers14092322
Muscarinic Receptors Associated with Cancer
Abstract
Cancer has been considered the pathology of the century and factors such as the environment may play an important etiological role. The ability of muscarinic agonists to stimulate growth and muscarinic receptor antagonists to inhibit tumor growth has been demonstrated for breast, melanoma, lung, gastric, colon, pancreatic, ovarian, prostate, and brain cancer. This work aimed to study the correlation between epidermal growth factor receptors and cholinergic muscarinic receptors, the survival differences adjusted by the stage clinical factor, and the association between gene expression and immune infiltration level in breast, lung, stomach, colon, liver, prostate, and glioblastoma human cancers. Thus, targeting cholinergic muscarinic receptors appears to be an attractive therapeutic alternative due to the complex signaling pathways involved.
Keywords: breast; cancer; colorectal; gastric; glioblastoma; liver; lung; muscarinic receptors; prostate.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

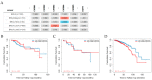


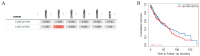

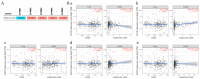
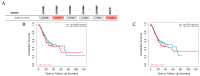



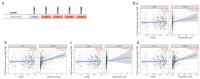

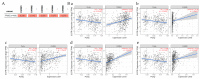


References
-
- International Agency for Research on Cancer; Lyon, France: 2020. p. 596.
-
- Gittelman M. The revolution re-visited: Clinical and genetics research paradigms and the productivity paradox in drug discovery. Res. Policy. 2016;45:1570–1585. doi: 10.1016/j.respol.2016.01.007. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

