Longitudinal Changes of the Ruminal Microbiota in Angus Beef Steers
- PMID: 35565493
- PMCID: PMC9102304
- DOI: 10.3390/ani12091066
Longitudinal Changes of the Ruminal Microbiota in Angus Beef Steers
Abstract
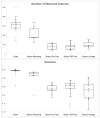
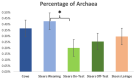
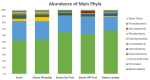
The ruminal microbiota of Angus cows and steers were characterized using 16s rRNA gene sequencing, and the expression of their metabolic pathways was predicted. Samples were collected on weaning day from the steers and the cows, and subsequently on three other occasions from the steers. Results showed that microbial richness, evenness, and diversity decreased (p < 0.001) in the rumen of the steers as they were weaned and transitioned to a high-concentrate feedlot diet. However, on the day of weaning, microbial evenness was similar to that observed in the rumen of cows (p = 0.12). The abundance of archaea was similar (p = 0.59) between the cows and steers at weaning, but it decreased (p = 0.04) in the rumen of steers after weaning, and remained stable (p ≥ 0.44) for the remainder of their lives. Likewise, no difference (p = 0.51) in the abundance of Bacteroidetes was detected between the cows and the calves on the day they were weaned, but the abundance of this phylum increased (p = 0.001) and remained stable after that. These results suggest that cows may have a strong influence on the composition, and help modulate the ruminal microbiota of young calves; however, following weaning, their ruminal microbiotas tend to differentiate from that state observed at earlier ages.
Keywords: calf; cow; dam; metabolic pathway; microbiome; rumen development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hungate R.E. The Rumen and Its Microbes. Academic Press; New York, NY, USA: 1966.
LinkOut - more resources
Full Text Sources

