Comprehensive Analysis of the Structure and Function of Peptide:N-Glycanase 1 and Relationship with Congenital Disorder of Deglycosylation
- PMID: 35565658
- PMCID: PMC9102325
- DOI: 10.3390/nu14091690
Comprehensive Analysis of the Structure and Function of Peptide:N-Glycanase 1 and Relationship with Congenital Disorder of Deglycosylation
Abstract
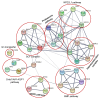
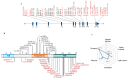
The cytosolic PNGase (peptide:N-glycanase), also known as peptide-N4-(N-acetyl-β-glucosaminyl)-asparagine amidase, is a well-conserved deglycosylation enzyme (EC 3.5.1.52) which catalyzes the non-lysosomal hydrolysis of an N(4)-(acetyl-β-d-glucosaminyl) asparagine residue (Asn, N) into a N-acetyl-β-d-glucosaminyl-amine and a peptide containing an aspartate residue (Asp, D). This enzyme (NGLY1) plays an essential role in the clearance of misfolded or unassembled glycoproteins through a process named ER-associated degradation (ERAD). Accumulating evidence also points out that NGLY1 deficiency can cause an autosomal recessive (AR) human genetic disorder associated with abnormal development and congenital disorder of deglycosylation. In addition, the loss of NGLY1 can affect multiple cellular pathways, including but not limited to NFE2L1 pathway, Creb1/Atf1-AQP pathway, BMP pathway, AMPK pathway, and SLC12A2 ion transporter, which might be the underlying reasons for a constellation of clinical phenotypes of NGLY1 deficiency. The current comprehensive review uncovers the NGLY1'ssdetailed structure and its important roles for participation in ERAD, involvement in CDDG and potential treatment for NGLY1 deficiency.
Keywords: ER associated degradation process; N-glycosylation; NFE2L1; NGLY1; congenital disorder of deglycosylation.
Conflict of interest statement
There were no competing interests between the authors and funding.
Figures



References
-
- Enns G.M., Shashi V., Bainbridge M., Gambello M.J., Zahir F.R., Bast T., Crimian R., Schoch K., Platt J., Cox R., et al. Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway. Genet. Med. 2014;16:751–758. doi: 10.1038/gim.2014.22. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

