Rapid Characterization of Bacterial Lipids with Ambient Ionization Mass Spectrometry for Species Differentiation
- PMID: 35566120
- PMCID: PMC9104219
- DOI: 10.3390/molecules27092772
Rapid Characterization of Bacterial Lipids with Ambient Ionization Mass Spectrometry for Species Differentiation
Abstract
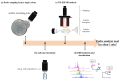
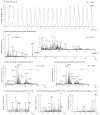
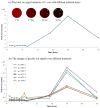
Ambient ionization mass spectrometry (AIMS) is both labor and time saving and has been proven to be useful for the rapid delineation of trace organic and biological compounds with minimal sample pretreatment. Herein, an analytical platform of probe sampling combined with a thermal desorption-electrospray ionization/mass spectrometry (TD-ESI/MS) and multivariate statistical analysis was developed to rapidly differentiate bacterial species based on the differences in their lipid profiles. For comparison, protein fingerprinting was also performed with matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) to distinguish these bacterial species. Ten bacterial species, including five Gram-negative and five Gram-positive bacteria, were cultured, and the lipids in the colonies were characterized with TD-ESI/MS. As sample pretreatment was unnecessary, the analysis of the lipids in a bacterial colony growing on a Petri dish was completed within 1 min. The TD-ESI/MS results were further performed by principal component analysis (PCA) and hierarchical cluster analysis (HCA) to assist the classification of the bacteria, and a low relative standard deviation (5.2%) of the total ion current was obtained from repeated analyses of the lipids in a single bacterial colony. The PCA and HCA results indicated that different bacterial species were successfully distinguished by the differences in their lipid profiles as validated by the differences in their protein profiles recorded from the MALDI-TOF analysis. In addition, real-time monitoring of the changes in the specific lipids of a colony with growth time was also achieved with probe sampling and TD-ESI/MS. The developed analytical platform is promising as a useful diagnostic tool by which to rapidly distinguish bacterial species in clinical practice.
Keywords: ambient ionization mass spectrometry; bacterial specie; lipid profile; multivariate statistical analysis; thermal desorption–electrospray ionization/mass spectrometry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Surface analysis of lipids by mass spectrometry: more than just imaging.Prog Lipid Res. 2013 Oct;52(4):329-53. doi: 10.1016/j.plipres.2013.04.005. Epub 2013 Apr 24. Prog Lipid Res. 2013. PMID: 23623802 Review.
-
Characteristics of glycation and glycation sites of lysozyme by matrix-assisted laser desorption/ionization time of flight/time-of-flight mass spectrometry and Liquid chromatography-electrospray ionization tandem mass spectrometry.Eur J Mass Spectrom (Chichester). 2014;20(4):327-36. Eur J Mass Spectrom (Chichester). 2014. PMID: 25420345
-
Comparative analysis of PCR-electrospray ionization/mass spectrometry (MS) and MALDI-TOF/MS for the identification of bacteria and yeast from positive blood culture bottles.Clin Chem. 2011 Jul;57(7):1057-67. doi: 10.1373/clinchem.2011.161968. Epub 2011 May 16. Clin Chem. 2011. PMID: 21576270 Free PMC article.
-
Bacterial identification by lipid profiling using liquid atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry.Clin Chem Lab Med. 2020 Jun 25;58(6):930-938. doi: 10.1515/cclm-2019-0908. Clin Chem Lab Med. 2020. PMID: 31851611
-
[Progress in applications of ambient ionization mass spectrometry for lipids identification].Se Pu. 2025 Jan;43(1):22-32. doi: 10.3724/SP.J.1123.2024.06007. Se Pu. 2025. PMID: 39722618 Free PMC article. Review. Chinese.
Cited by
-
Role of Oxylipins in the Inflammatory-Related Diseases NAFLD, Obesity, and Type 2 Diabetes.Metabolites. 2022 Dec 9;12(12):1238. doi: 10.3390/metabo12121238. Metabolites. 2022. PMID: 36557276 Free PMC article. Review.
-
Rapid Screening of Etomidate and Its Analogs in Seized e-Liquids Using Thermal Desorption Electrospray Ionization Coupled with Triple Quadrupole Mass Spectrometry.Toxics. 2024 Dec 5;12(12):884. doi: 10.3390/toxics12120884. Toxics. 2024. PMID: 39771099 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

