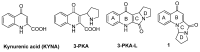Synthesis and Investigation of the G-Quadruplex Binding Properties of Kynurenic Acid Derivatives with a Dihydroimidazoquinoline-3,5-dione Core
- PMID: 35566141
- PMCID: PMC9103425
- DOI: 10.3390/molecules27092791
Synthesis and Investigation of the G-Quadruplex Binding Properties of Kynurenic Acid Derivatives with a Dihydroimidazoquinoline-3,5-dione Core
Abstract
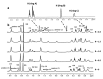
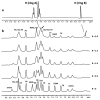
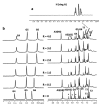
G-quadruplexes are secondary structures originating from nucleic acid regions rich in guanines, which are well known for their involvement in gene transcription and regulation and DNA damage repair. In recent studies from our group, kynurenic acid (KYNA) derivative 1 was synthesized and found to share the structural features typical of G-quadruplex binders. Herein, structural modifications were conducted on this scaffold in order to assist the binding with a G-quadruplex, by introducing charged hydrophilic groups. The antiproliferative activity of the new analogues was evaluated on an IGROV-1 human ovarian cancer cell line, and the most active compound, compound 9, was analyzed with NMR spectrometry in order to investigate its binding mode with DNA. The results indicated that a weak, non-specific interaction was set with duplex nucleotides; on the other hand, titration in the presence of a G-quadruplex from human telomere d(TTAGGGT)4 showed a stable, although not strong, interaction at the 3'-end of the nucleotidic sequence, efficiently assisted by salt bridges between the quaternary nitrogen and the external phosphate groups. Overall, this work can be considered a platform for the development of a new class of potential G-quadruplex stabilizing molecules, confirming the crucial role of a planar system and the ability of charged nitrogen-containing groups to facilitate the binding to G-quadruplex grooves and loops.
Keywords: DNA; G-quadruplex; KYNA; cytotoxicity; kynurenic acid; stabilization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures