Synthetic Receptors Based on Abiotic Cyclo(pseudo)peptides
- PMID: 35566168
- PMCID: PMC9103335
- DOI: 10.3390/molecules27092821
Synthetic Receptors Based on Abiotic Cyclo(pseudo)peptides
Abstract
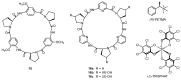
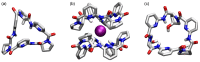
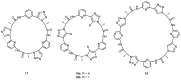
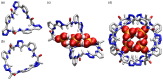
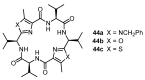
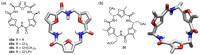
Work on the use of cyclic peptides or pseudopeptides as synthetic receptors started even before the field of supramolecular chemistry was firmly established. Research initially focused on the development of synthetic ionophores and involved the use of macrocycles with a repeating sequence of subunits along the ring to facilitate the correlation between structure, conformation, and binding properties. Later, nonnatural amino acids as building blocks were also considered. With growing research in this area, cyclopeptides and related macrocycles developed into an important and structurally diverse receptor family. This review provides an overview of these developments, starting from the early years. The presented systems are classified according to characteristic structural elements present along the ring. Wherever possible, structural aspects are correlated with binding properties to illustrate how natural or nonnatural amino acids affect binding properties.
Keywords: amino acids; cyclic pseudopeptides; cyclopeptides; molecular recognition; supramolecular chemistry; synthetic receptors.
Conflict of interest statement
The author declares no conflict of interest.
Figures

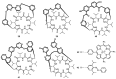
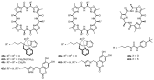

Similar articles
-
Anion Recognition in Aqueous Media by Cyclopeptides and Other Synthetic Receptors.Acc Chem Res. 2017 Nov 21;50(11):2870-2878. doi: 10.1021/acs.accounts.7b00458. Epub 2017 Nov 10. Acc Chem Res. 2017. PMID: 29125287 Review.
-
Selective Recognition of Amino Acids and Peptides by Small Supramolecular Receptors.Molecules. 2020 Dec 28;26(1):106. doi: 10.3390/molecules26010106. Molecules. 2020. PMID: 33379401 Free PMC article. Review.
-
Cu(II) coordination chemistry of patellamide derivatives: possible biological functions of cyclic pseudopeptides.Chemistry. 2012 Feb 27;18(9):2578-90. doi: 10.1002/chem.201101975. Epub 2012 Jan 20. Chemistry. 2012. PMID: 22266951
-
Selectivity Modulation and Structure of α/aza-β3 Cyclic Antimicrobial Peptides.Chemistry. 2018 Apr 20;24(23):6191-6201. doi: 10.1002/chem.201800152. Epub 2018 Mar 26. Chemistry. 2018. PMID: 29411917
-
Carbohydrates in peptide and protein design.Biopolymers. 2005;80(6):747-61. doi: 10.1002/bip.20300. Biopolymers. 2005. PMID: 15929028 Review.
Cited by
-
Anion-Binding Properties of Short Linear Homopeptides.Int J Mol Sci. 2024 May 11;25(10):5235. doi: 10.3390/ijms25105235. Int J Mol Sci. 2024. PMID: 38791275 Free PMC article.
-
Structure-Activity Relationship of Synthetic Linear KTS-Peptides Containing Meta-Aminobenzoic Acid as Antagonists of α1β1 Integrin with Anti-Angiogenic and Melanoma Anti-Tumor Activities.Pharmaceuticals (Basel). 2024 Apr 24;17(5):549. doi: 10.3390/ph17050549. Pharmaceuticals (Basel). 2024. PMID: 38794120 Free PMC article.
References
-
- Deber C.M., Madison V., Blout E.R. Why cyclic peptides—Complementary approaches to conformations. Acc. Chem. Res. 1976;9:106–113. doi: 10.1021/ar50099a005. - DOI
-
- Lehn J.-M. Supramolecular Chemistry. Wiley-VCH; Weinheim, Germany: 1995.
-
- Kubik S. In: Artificial Receptors for Chemical Sensors. Mirsky V.M., Yatsimirsky A.K., editors. Wiley-VCH; Weinheim, Germany: 2010. pp. 135–167. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

