New Insights into the Genomic Structure of Avena L.: Comparison of the Divergence of A-Genome and One C-Genome Oat Species
- PMID: 35567104
- PMCID: PMC9102028
- DOI: 10.3390/plants11091103
New Insights into the Genomic Structure of Avena L.: Comparison of the Divergence of A-Genome and One C-Genome Oat Species
Abstract
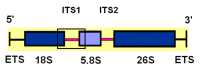
We used next-generation sequencing analysis of the 3′-part of 18S rDNA, ITS1, and a 5′-part of the 5.8S rDNA region to understand genetic variation among seven diploid A-genome Avena species. We used 4−49 accessions per species that represented the As genome (A. atlantica, A. hirtula, and wiestii), Ac genome (A. canariensis), Ad genome (A. damascena), Al genome (A. longiglumis), and Ap genome (A. prostrata). We also took into our analysis one C-genome species, A. clauda, which previously was found to be related to A-genome species. The sequences of 169 accessions revealed 156 haplotypes of which seven haplotypes were shared by two to five species. We found 16 ribotypes that consisted of a unique sequence with a characteristic pattern of single nucleotide polymorphisms and deletions. The number of ribotypes per species varied from one in A. longiglumis to four in A. wiestii. Although most ribotypes were species-specific, we found two ribotypes shared by three species (one for A. damascena, A. hirtula, and A. wiestii, and the second for A. longiglumis, A. atlantica, and A. wiestii), and a third ribotype shared between A. atlantica and A. wiestii. A characteristic feature of the A. clauda ribotype, a diploid C-genome species, is that two different families of ribotypes have been found in this species. Some of these ribotypes are characteristic of Cc-genome species, whereas others are closely related to As-genome ribotypes. This means that A. clauda can be a hybrid between As- and C-genome oats.
Keywords: grasses; hybridization; molecular phylogeny; next-generation sequencing; rDNA; rRNA genes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Soreng R.J., Peterson P.M., Romaschenko K., Davidse G., Judziewicz E.J., Zuloaga F.O., Filgueiras T.S., Morrone O. A worldwide phylogenetic classification of the Poaceae (Gramineae) J. Syst. Evol. 2015;53:117–137. doi: 10.1111/jse.12150. - DOI
-
- Saarela J.M., Bull R.D., Paradis M.J., Ebata S.N., Peterson P.M., Soreng R.J., Paszko B. Molecular phylogenetics of cool-season grasses in the subtribes Agrostidinae, Anthoxanthinae, Aveninae, Brizinae, Calothecinae, Koeleriinae and Phalaridinae (Poaceae, Pooideae, Poeae, Poeae chloroplast group 1) PhytoKeys. 2017;87:1–139. doi: 10.3897/phytokeys.87.12774. - DOI - PMC - PubMed
-
- Loskutov I.G., Rines H.W. Avena. In: Kole C., editor. Wild Crop Relatives: Genomic and Breeding Resources. Springer; Berlin/Heidelberg, Germany: 2011. pp. 109–183. Chapter 3.
-
- Rajhathy T., Thomas H. Cytogenetics of Oats (Avena L.) Miscellaneous Publications of the Genetics Society of Canada 2. Ontario (Canada) Genetics Society of Canada; Ottawa, ON, Canada: 1974. 90p
-
- Fominaya A., Vega C., Ferrer E. Giemsa C-banded karyotypes of Avena species. Genome. 1988;30:627–632. doi: 10.1139/g88-106. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

