Transcriptomic Analysis Elaborates the Resistance Mechanism of Grapevine Rootstocks against Salt Stress
- PMID: 35567166
- PMCID: PMC9103662
- DOI: 10.3390/plants11091167
Transcriptomic Analysis Elaborates the Resistance Mechanism of Grapevine Rootstocks against Salt Stress
Abstract
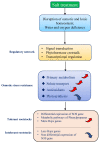
Grapes are subject to a wide range of climatic conditions during their life cycle, but the use of rootstocks can effectively ameliorate the effects of abiotic stress. However, the tolerance mechanism of different grape rootstock varieties varies under various stresses, and systematic research on this aspect is limited. On the basis of previous research, transcriptome sequencing was performed on three tolerant grape rootstock varieties (3309C, 520A, 1103P) and three intolerant grape rootstock varieties (5BB, 101-14, Beta). In total, 56,478,468 clean reads were obtained. One hundred and ten genes only existed in all combinations during P1 with a downregulated trend, and 178 genes existed only in P1 of tolerant grape rootstock varieties. Salt treatment firstly affected the photosynthesis of leaves, and tolerant varieties weakened or even eliminated this effect through their own mechanisms in the later stage. Tolerant varieties mobilized a large number of MFs during the P2 stage, such as hydrolase activity, carboxypeptidase activity, and dioxygenase activity. Carbon metabolism was significantly enriched in P1, while circadian rhythm and flavonoid biosynthesis were only enriched in tolerant varieties. In the intolerant varieties, photosynthesis-related pathways were always the most significantly enriched. There were large differences in the gene expression of the main signal pathways related to salt stress in different varieties. Salt stress affected the expression of genes related to plant abiotic stress, biotic stress, transcription factors, hormones, and secondary metabolism. Tolerant varieties mobilized more bHLH, WRKY, and MYB transcription factors to respond to salt stress than intolerant varieties. In the tolerant rootstocks, SOS was co-expressed. Among these, SOS1 and SOS2 were upregulated, and the SOS3 and SOS5 components were downregulated. The genes of heat shock proteins and the phenylalanine pathway were upregulated in the tolerant varieties. These findings outline a tolerance mechanism model for rootstocks for coping with osmotic stress, providing important information for improving the resistance of grapes under global climate change.
Keywords: grape; rootstock; salt stress; signal transduction.
Conflict of interest statement
The authors declare no competing interest.
Figures






References
-
- Keller M. Managing grapevines to optimise fruit development in a challenging environment: A climate change primer for viticulturists. Aust. J. Grape Wine Res. 2010;16:56–69. doi: 10.1111/j.1755-0238.2009.00077.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

