Proteomic Investigation of Molecular Mechanisms in Response to PEG-Induced Drought Stress in Soybean Roots
- PMID: 35567174
- PMCID: PMC9100407
- DOI: 10.3390/plants11091173
Proteomic Investigation of Molecular Mechanisms in Response to PEG-Induced Drought Stress in Soybean Roots
Abstract
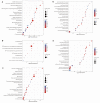
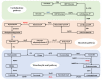
Roots are generally the critical drought sensors, but little is known about their molecular response to drought stress. We used the drought-tolerant soybean variety 'Jiyu 47' to investigate the differentially expressed proteins (DEPs) in soybean roots during the seedling stage based on the tandem mass tag (TMT) proteomics analysis. Various expression patterns were observed in a total of six physiological parameters. A total of 468 DEPs (144 up-regulated and 324 down-regulated) among a total of 8687 proteins were identified in response to drought stress in 24 h. The expression of DEPs was further validated based on quantitative real-time PCR of a total of five genes (i.e., GmGSH, GmGST1, GmGST2 k GmCAT, and Gm6PGD) involved in the glutathione biosynthesis. Results of enrichment analyses revealed a coordinated expression pattern of proteins involved in various cellular metabolisms responding to drought stress in soybean roots. Our results showed that drought stress caused significant alterations in the expression of proteins involved in several metabolic pathways in soybean roots, including carbohydrate metabolism, metabolism of the osmotic regulation substances, and antioxidant defense system (i.e., the glutathione metabolism). Increased production of reduced glutathione (GSH) enhanced the prevention of the damage caused by reactive oxygen species and the tolerance of the abiotic stress. The glutathione metabolism played a key role in modifying the antioxidant defense system in response to drought stress in soybean roots. Our proteomic study suggested that the soybean plants responded to drought stress by coordinating their protein expression during the vegetative stage, providing novel insights into the molecular mechanisms regulating the response to abiotic stress in plants.
Keywords: antioxidant; drought stress; enrichment analysis; gene ontology; glutathione; physiological response; proteomics; soybean root; tandem mass tag.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Tandem mass tag-based (TMT) quantitative proteomics analysis reveals the response of fine roots to drought stress in cotton (Gossypium hirsutum L.).BMC Plant Biol. 2020 Jul 11;20(1):328. doi: 10.1186/s12870-020-02531-z. BMC Plant Biol. 2020. PMID: 32652934 Free PMC article.
-
Comparative Proteomics Analysis of the Seedling Root Response of Drought-sensitive and Drought-tolerant Maize Varieties to Drought Stress.Int J Mol Sci. 2019 Jun 7;20(11):2793. doi: 10.3390/ijms20112793. Int J Mol Sci. 2019. PMID: 31181633 Free PMC article.
-
Physiological and Differential Proteomic Analyses of Imitation Drought Stress Response in Sorghum bicolor Root at the Seedling Stage.Int J Mol Sci. 2020 Dec 1;21(23):9174. doi: 10.3390/ijms21239174. Int J Mol Sci. 2020. PMID: 33271965 Free PMC article.
-
Proteomic approaches to uncover the flooding and drought stress response mechanisms in soybean.J Proteomics. 2018 Feb 10;172:201-215. doi: 10.1016/j.jprot.2017.11.006. Epub 2017 Nov 11. J Proteomics. 2018. PMID: 29133124 Review.
-
Potentiality of Soybean Proteomics in Untying the Mechanism of Flood and Drought Stress Tolerance.Proteomes. 2014 Mar 7;2(1):107-127. doi: 10.3390/proteomes2010107. Proteomes. 2014. PMID: 28250373 Free PMC article. Review.
Cited by
-
Monitoring Drought Stress in Common Bean Using Chlorophyll Fluorescence and Multispectral Imaging.Plants (Basel). 2023 Mar 21;12(6):1386. doi: 10.3390/plants12061386. Plants (Basel). 2023. PMID: 36987074 Free PMC article.
-
Comprehensive Proteomic Analysis of Common Bean (Phaseolus vulgaris L.) Seeds Reveal Shared and Unique Proteins Involved in Terminal Drought Stress Response in Tolerant and Sensitive Genotypes.Biomolecules. 2024 Jan 15;14(1):109. doi: 10.3390/biom14010109. Biomolecules. 2024. PMID: 38254709 Free PMC article.
-
Transcriptomics-proteomics analysis reveals StCOMT1 regulates drought, alkali and combined stresses in potato.Plant Cell Rep. 2025 Apr 29;44(5):109. doi: 10.1007/s00299-025-03496-9. Plant Cell Rep. 2025. PMID: 40299051
-
Quantitative proteomic analysis based on TMT reveals different responses of Haloxylon ammodendron and Haloxylon persicum to long-term drought.BMC Plant Biol. 2025 Apr 15;25(1):480. doi: 10.1186/s12870-025-06513-x. BMC Plant Biol. 2025. PMID: 40234745 Free PMC article.
-
Integrated transcriptomic, proteomic and metabolomic analyses revealing the roles of amino acid and sucrose metabolism in augmenting drought tolerance in Agropyron mongolicum.Front Plant Sci. 2024 Dec 16;15:1515944. doi: 10.3389/fpls.2024.1515944. eCollection 2024. Front Plant Sci. 2024. PMID: 39741683 Free PMC article.
References
-
- Agrawal L., Gupta S., Mishra S.K., Pandey G., Kumar S., Chauhan P.S., Chakrabarty D., Nautiyal C.S. Elucidation of Complex Nature of PEG Induced Drought-Stress Response in Rice Root Using Comparative Proteomics Approach. Front. Plant Sci. 2016;7:1466. doi: 10.3389/fpls.2016.01466. - DOI - PMC - PubMed
-
- Zhang S.-H., Xu X.-F., Sun Y.-M., Zhang J.-L., Li C.-Z. Influence of drought hardening on the resistance physiology of potato seedlings under drought stress. J. Integr. Agric. 2018;17:336–347. doi: 10.1016/S2095-3119(17)61758-1. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

