Adaptive Potential of Epigenetic Switching During Adaptation to Fluctuating Environments
- PMID: 35567483
- PMCID: PMC9113428
- DOI: 10.1093/gbe/evac065
Adaptive Potential of Epigenetic Switching During Adaptation to Fluctuating Environments
Abstract
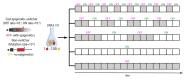
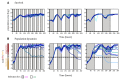
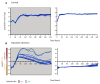
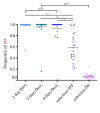
Epigenetic regulation of gene expression allows for the emergence of distinct phenotypic states within the clonal population. Due to the instability of epigenetic inheritance, these phenotypes can intergenerationally switch between states in a stochastic manner. Theoretical studies of evolutionary dynamics predict that the phenotypic heterogeneity enabled by this rapid epigenetic switching between gene expression states would be favored under fluctuating environmental conditions, whereas genetic mutations, as a form of stable inheritance system, would be favored under a stable environment. To test this prediction, we engineered switcher and non-switcher yeast strains, in which the uracil biosynthesis gene URA3 is either continually expressed or switched on and off at two different rates (slow and fast switchers). Competitions between clones with an epigenetically controlled URA3 and clones without switching ability (SIR3 knockout) show that the switchers are favored in fluctuating environments. This occurs in conditions where the environments fluctuate at similar rates to the rate of switching. However, in stable environments, but also in environments with fluctuation frequency higher than the rate of switching, we observed that genetic changes dominated. Remarkably, epigenetic clones with a high, but not with a low, rate of switching can coexist with non-switchers even in a constant environment. Our study offers an experimental proof of concept that helps defining conditions of environmental fluctuation under which epigenetic switching provides an advantage.
Keywords: adaptation; epigenetic switching; fluctuating environments; mutations.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Epigenetic switching as a strategy for quick adaptation while attenuating biochemical noise.PLoS Comput Biol. 2019 Oct 28;15(10):e1007364. doi: 10.1371/journal.pcbi.1007364. eCollection 2019 Oct. PLoS Comput Biol. 2019. PMID: 31658246 Free PMC article.
-
Stochastic switching as a survival strategy in fluctuating environments.Nat Genet. 2008 Apr;40(4):471-5. doi: 10.1038/ng.110. Epub 2008 Mar 23. Nat Genet. 2008. PMID: 18362885
-
Evolution of a bistable genetic system in fluctuating and nonfluctuating environments.Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2322371121. doi: 10.1073/pnas.2322371121. Epub 2024 Aug 30. Proc Natl Acad Sci U S A. 2024. PMID: 39213178 Free PMC article.
-
Empirical evidence for epigenetic inheritance driving evolutionary adaptation.Philos Trans R Soc Lond B Biol Sci. 2021 Jun 7;376(1826):20200121. doi: 10.1098/rstb.2020.0121. Epub 2021 Apr 19. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 33866813 Free PMC article. Review.
-
It's not magic - Hsp90 and its effects on genetic and epigenetic variation.Semin Cell Dev Biol. 2019 Apr;88:21-35. doi: 10.1016/j.semcdb.2018.05.015. Epub 2018 Jun 6. Semin Cell Dev Biol. 2019. PMID: 29807130 Free PMC article. Review.
Cited by
-
Environmental memory alters the fitness effects of adaptive mutations in fluctuating environments.Nat Ecol Evol. 2024 Sep;8(9):1760-1775. doi: 10.1038/s41559-024-02475-9. Epub 2024 Jul 17. Nat Ecol Evol. 2024. PMID: 39020024 Free PMC article.
-
Transgenerational epigenetic inheritance increases trait variation but is not adaptive.Evolution. 2025 Jun 14;79(6):1033-1043. doi: 10.1093/evolut/qpaf050. Evolution. 2025. PMID: 40065197 Free PMC article.
-
Transgenerational epigenetic inheritance increases trait variation but is not adaptive.bioRxiv [Preprint]. 2024 Apr 20:2024.04.15.589575. doi: 10.1101/2024.04.15.589575. bioRxiv. 2024. Update in: Evolution. 2025 Jun 14;79(6):1033-1043. doi: 10.1093/evolut/qpaf050. PMID: 38659883 Free PMC article. Updated. Preprint.
-
Strong environmental memory revealed by experimental evolution in static and fluctuating environments.bioRxiv [Preprint]. 2023 Sep 21:2023.09.14.557739. doi: 10.1101/2023.09.14.557739. bioRxiv. 2023. Update in: Nat Ecol Evol. 2024 Sep;8(9):1760-1775. doi: 10.1038/s41559-024-02475-9. PMID: 37745585 Free PMC article. Updated. Preprint.
-
Environmental Adaptation of Genetically Uniform Organisms with the Help of Epigenetic Mechanisms-An Insightful Perspective on Ecoepigenetics.Epigenomes. 2022 Dec 26;7(1):1. doi: 10.3390/epigenomes7010001. Epigenomes. 2022. PMID: 36648862 Free PMC article. Review.
References
-
- Acar M, Mettetal JT, Van Oudenaarden A. 2008. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 40:471–475. - PubMed
-
- Adrian-Kalchhauser I, et al. 2020. Understanding ‘non-genetic’ inheritance: insights from molecular-evolutionary crosstalk. Trends Ecol Evol. 35(12):1078–1089. - PubMed
-
- Aparicio OM, Billington BL, Gottschling DE. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279–1287. - PubMed
-
- Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB. 2009. Experimental evolution of bet hedging. Nature 462(7269):90–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

