Multiomics identifies the link between intratumor steatosis and the exhausted tumor immune microenvironment in hepatocellular carcinoma
- PMID: 35567547
- PMCID: PMC9970024
- DOI: 10.1002/hep.32573
Multiomics identifies the link between intratumor steatosis and the exhausted tumor immune microenvironment in hepatocellular carcinoma
Abstract
Background and aims: Immunotherapy has become the standard-of-care treatment for hepatocellular carcinoma (HCC), but its efficacy remains limited. To identify immunotherapy-susceptible HCC, we profiled the molecular abnormalities and tumor immune microenvironment (TIME) of rapidly increasing nonviral HCC.
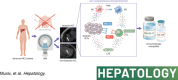
Approaches and results: We performed RNA-seq of tumor tissues in 113 patients with nonviral HCC and cancer genome sequencing of 69 genes with recurrent genetic alterations reported in HCC. Unsupervised hierarchical clustering classified nonviral HCCs into three molecular classes (Class I, II, III), which stratified patient prognosis. Class I, with the poorest prognosis, was associated with TP53 mutations, whereas class III, with the best prognosis, was associated with cadherin-associated protein beta 1 (CTNNB1) mutations. Thirty-eight percent of nonviral HCC was defined as an immune class characterized by a high frequency of intratumoral steatosis and a low frequency of CTNNB1 mutations. Steatotic HCC, which accounts for 23% of nonviral HCC cases, presented an immune-enriched but immune-exhausted TIME characterized by T cell exhaustion, M2 macrophage and cancer-associated fibroblast (CAF) infiltration, high PD-L1 expression, and TGF-β signaling activation. Spatial transcriptome analysis suggested that M2 macrophages and CAFs may be in close proximity to exhausted CD8+ T cells in steatotic HCC. An in vitro study showed that palmitic acid-induced lipid accumulation in HCC cells upregulated PD-L1 expression and promoted immunosuppressive phenotypes of cocultured macrophages and fibroblasts. Patients with steatotic HCC, confirmed by chemical-shift MR imaging, had significantly longer PFS with combined immunotherapy using anti-PD-L1 and anti-VEGF antibodies.
Conclusions: Multiomics stratified nonviral HCCs according to prognosis or TIME. We identified the link between intratumoral steatosis and immune-exhausted immunotherapy-susceptible TIME.
Copyright © 2022 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
Hayato Hikita, Takahiro Kodama and Tetsuo Takehara are on the speakers' bureau for Chugai Pharmaceutical Co., Ltd.
Figures






Comment in
-
Letter to the editor: Is the intratumor steatosis a novel biomarker for predicting the efficacy of immunotherapy in hepatocellular carcinoma?Hepatology. 2022 Dec;76(6):E131-E132. doi: 10.1002/hep.32616. Epub 2022 Jun 22. Hepatology. 2022. PMID: 35702874 No abstract available.
-
Imaging for better responses to immunotherapy in hepatocellular carcinoma.Hepatology. 2023 Jan 1;77(1):6-9. doi: 10.1002/hep.32627. Epub 2022 Jul 10. Hepatology. 2023. PMID: 35727179 Free PMC article. No abstract available.
-
Letter to the editor: Using intratumoral steatosis to predict the efficacy of combination therapy in nonviral hepatocellular carcinoma.Hepatology. 2022 Dec;76(6):E135-E136. doi: 10.1002/hep.32628. Epub 2022 Jul 4. Hepatology. 2022. PMID: 35751482 No abstract available.
-
Lipids link immune suppression to effective immunotherapy in steatotic hepatocellular carcinoma.Ann Transl Med. 2023 Mar 15;11(5):226. doi: 10.21037/atm-22-6548. Epub 2023 Jan 16. Ann Transl Med. 2023. PMID: 37007550 Free PMC article. No abstract available.
References
-
- Paradis V. Histopathology of hepatocellular carcinoma. Recent Results Cancer Res. 2013;190:21–32. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

