Identifying the individual metabolic abnormities from a systemic perspective using whole-body PET imaging
- PMID: 35567627
- PMCID: PMC9106794
- DOI: 10.1007/s00259-022-05832-7
Identifying the individual metabolic abnormities from a systemic perspective using whole-body PET imaging
Abstract
Introduction: Distinct physiological states arise from complex interactions among the various organs present in the human body. PET is a non-invasive modality with numerous successful applications in oncology, neurology, and cardiology. However, while PET imaging has been applied extensively in detecting focal lesions or diseases, its potential in detecting systemic abnormalities is seldom explored, mostly because total-body imaging was not possible until recently.
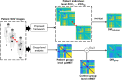
Methods: In this context, the present study proposes a framework capable of constructing an individual metabolic abnormality network using a subject's whole-body 18F-FDG SUV image and a normal control database. The developed framework was evaluated in the patients with lung cancer, the one discharged after suffering from Covid-19 disease, and the one that had gastrointestinal bleeding with the underlying cause unknown.
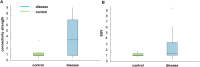
Results: The framework could successfully capture the deviation of these patients from healthy subjects at the level of both system and organ. The strength of the altered network edges revealed the abnormal metabolic connection between organs. The overall deviation of the network nodes was observed to be highly correlated to the organ SUV measures. Therefore, the molecular connectivity of glucose metabolism was characterized at a single subject level.
Conclusion: The proposed framework represents a significant step toward the use of PET imaging for identifying metabolic dysfunction from a systemic perspective. A better understanding of the underlying biological mechanisms and the physiological interpretation of the interregional connections identified in the present study warrant further research.
Keywords: Metabolic abnormality; Network analysis; Systemic disease; Whole-body PET.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

