Epithelial cells adapt to curvature induction via transient active osmotic swelling
- PMID: 35568030
- PMCID: PMC9165930
- DOI: 10.1016/j.devcel.2022.04.017
Epithelial cells adapt to curvature induction via transient active osmotic swelling
Abstract
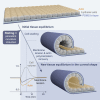
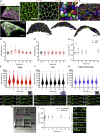
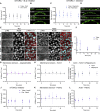
Generation of tissue curvature is essential to morphogenesis. However, how cells adapt to changing curvature is still unknown because tools to dynamically control curvature in vitro are lacking. Here, we developed self-rolling substrates to study how flat epithelial cell monolayers adapt to a rapid anisotropic change of curvature. We show that the primary response is an active and transient osmotic swelling of cells. This cell volume increase is not observed on inducible wrinkled substrates, where concave and convex regions alternate each other over short distances; and this finding identifies swelling as a collective response to changes of curvature with a persistent sign over large distances. It is triggered by a drop in membrane tension and actin depolymerization, which is perceived by cells as a hypertonic shock. Osmotic swelling restores tension while actin reorganizes, probably to comply with curvature. Thus, epithelia are unique materials that transiently and actively swell while adapting to large curvature induction.
Keywords: cell volume; curvature; epithelial mechanics; mTORC2; membrane tension.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Baptista D., Teixeira L., van Blitterswijk C., Giselbrecht S., Truckenmüller R. Overlooked? Underestimated? Effects of substrate curvature on cell behavior. Trends Biotechnol. 2019;37:838–854. - PubMed
-
- Baughman R.H., Stafström S., Cui C., Dantas S.O. Materials with negative compressibilities in one or more dimensions. Science. 1998;279:1522–1524. - PubMed
-
- Blonski S., Aureille J., Badawi S., Zaremba D., Pernet L., Grichine A., Fraboulet S., Korczyk P.M., Recho P., Guilluy C., et al. Direction of epithelial folding defines impact of mechanical forces on epithelial state. Dev. Cell. 2021;56:3222–3234.e6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

