Protective prototype-Beta and Delta-Omicron chimeric RBD-dimer vaccines against SARS-CoV-2
- PMID: 35568034
- PMCID: PMC9042943
- DOI: 10.1016/j.cell.2022.04.029
Protective prototype-Beta and Delta-Omicron chimeric RBD-dimer vaccines against SARS-CoV-2
Abstract
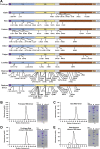
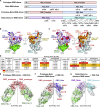
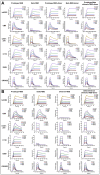
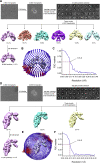
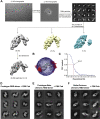
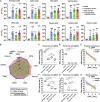
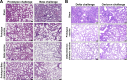
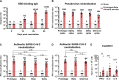
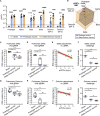
Breakthrough infections by SARS-CoV-2 variants become the global challenge for pandemic control. Previously, we developed the protein subunit vaccine ZF2001 based on the dimeric receptor-binding domain (RBD) of prototype SARS-CoV-2. Here, we developed a chimeric RBD-dimer vaccine approach to adapt SARS-CoV-2 variants. A prototype-Beta chimeric RBD-dimer was first designed to adapt the resistant Beta variant. Compared with its homotypic forms, the chimeric vaccine elicited broader sera neutralization of variants and conferred better protection in mice. The protection of the chimeric vaccine was further verified in macaques. This approach was generalized to develop Delta-Omicron chimeric RBD-dimer to adapt the currently prevalent variants. Again, the chimeric vaccine elicited broader sera neutralization of SARS-CoV-2 variants and conferred better protection against challenge by either Delta or Omicron SARS-CoV-2 in mice. The chimeric approach is applicable for rapid updating of immunogens, and our data supported the use of variant-adapted multivalent vaccine against circulating and emerging variants.
Keywords: COVID19 vaccine; Delta variant; Omicron variant; RBD; SARS-CoV-2; VOC; immunogen structure; receptor-binding domain; vaccine protection; variant of concern.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Kun Xu, Y.A., L.D., and G.F.G. are listed in the patent as the inventors of the prototype RBD-dimer as coronavirus vaccines. Kun Xu, P.G., Z.Z., Y.A., L.D., and G.F.G. are listed in the patent as the inventors of chimeric prototype-Beta RBD-dimer as coronavirus vaccines. Kun Xu, T.Z., L.D., and G.F.G. are listed in the patent as the inventors of chimeric Delta-Omicron RBD-dimer as coronavirus vaccine. All other authors declare no competing interests.
Figures


References
-
- Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M.J., Lai L., Edara V.V., et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594:253–258. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

