A family of conserved bacterial virulence factors dampens interferon responses by blocking calcium signaling
- PMID: 35568036
- PMCID: PMC9596379
- DOI: 10.1016/j.cell.2022.04.028
A family of conserved bacterial virulence factors dampens interferon responses by blocking calcium signaling
Abstract
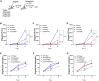
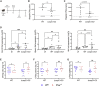
Interferons (IFNs) induce an antimicrobial state, protecting tissues from infection. Many viruses inhibit IFN signaling, but whether bacterial pathogens evade IFN responses remains unclear. Here, we demonstrate that the Shigella OspC family of type-III-secreted effectors blocks IFN signaling independently of its cell death inhibitory activity. Rather, IFN inhibition was mediated by the binding of OspC1 and OspC3 to the Ca2+ sensor calmodulin (CaM), blocking CaM kinase II and downstream JAK/STAT signaling. The growth of Shigella lacking OspC1 and OspC3 was attenuated in epithelial cells and in a murine model of infection. This phenotype was rescued in both models by the depletion of IFN receptors. OspC homologs conserved in additional pathogens not only bound CaM but also inhibited IFN, suggesting a widespread virulence strategy. These findings reveal a conserved but previously undescribed molecular mechanism of IFN inhibition and demonstrate the critical role of Ca2+ and IFN targeting in bacterial pathogenesis.
Keywords: Ca(2+); CaMKII; ISG; JAK/STAT; OspC1; OspC3; Shigella; T3SS; calcium; calmodulin; host-pathogen interactions; interferons.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures










Comment in
-
Shigella "Osp"pression of innate immunity.Cell. 2022 Jun 23;185(13):2205-2207. doi: 10.1016/j.cell.2022.05.027. Cell. 2022. PMID: 35750030
References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Anderson M.C., Vonaesch P., Saffarian A., Marteyn B.S., Sansonetti P.J. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe. 2017;21:769–776.e3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

