Mouse Chd4-NURD is required for neonatal spermatogonia survival and normal gonad development
- PMID: 35568926
- PMCID: PMC9107693
- DOI: 10.1186/s13072-022-00448-5
Mouse Chd4-NURD is required for neonatal spermatogonia survival and normal gonad development
Abstract
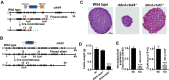
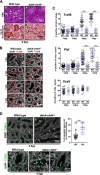
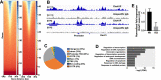
Testis development and sustained germ cell production in adults rely on the establishment and maintenance of spermatogonia stem cells and their proper differentiation into spermatocytes. Chromatin remodeling complexes regulate critical processes during gamete development by restricting or promoting accessibility of DNA repair and gene expression machineries to the chromatin. Here, we investigated the role of Chd4 and Chd3 catalytic subunits of the NURD complex during spermatogenesis. Germ cell-specific deletion of chd4 early in gametogenesis, but not chd3, resulted in arrested early gamete development due to failed cell survival of neonate undifferentiated spermatogonia stem cell population. Candidate assessment revealed that Chd4 controls expression of dmrt1 and its downstream target plzf, both described as prominent regulators of spermatogonia stem cell maintenance. Our results show the requirement of Chd4 in mammalian gametogenesis pointing to functions in gene expression early in the process.
Keywords: Chromatin remodeling; Mouse gametogenesis; NURD; Spermatogonia.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

