Genomic profiling of sporadic multiple meningiomas
- PMID: 35568945
- PMCID: PMC9107270
- DOI: 10.1186/s12920-022-01258-0
Genomic profiling of sporadic multiple meningiomas
Erratum in
-
Correction: Genomic profiling of sporadic multiple meningiomas.BMC Med Genomics. 2022 Jun 13;15(1):131. doi: 10.1186/s12920-022-01273-1. BMC Med Genomics. 2022. PMID: 35698142 Free PMC article. No abstract available.
Abstract
Background: Multiple meningiomas (MMs) rarely occur sporadically. It is unclear whether each individual tumor in a single patient behaves similarly. Moreover, the molecular mechanisms underlying the formation of sporadic MMs and clonal formation etiology of these tumors are poorly understood.
Methods: Patients with spatially separated MMs without prior radiation exposure or a family history who underwent surgical resection of at least two meningiomas were included. Unbiased, comprehensive next generation sequencing was performed, and relevant clinical data was analyzed.
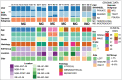
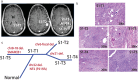
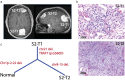
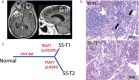
Results: Fifteen meningiomas and one dural specimen from six patients were included. The majority of tumors (12/15) were WHO Grade I; one patient had bilateral MMs, one of which was Grade II, while the other was Grade I. We found 11/15 of our cohort specimens were of NF2-loss subtype. Meningiomas from 5/6 patients had a monoclonal origin, with the tumor from the remaining patient showing evidence for independent clonal formation. We identified a novel case of non-NF2 mutant MM with monoclonal etiology. MMs due to a monoclonal origin did not always display a homogenous genomic profile, but rather exhibited heterogeneity due to branching evolution.
Conclusions: Both NF2-loss and non-NF2 driven MMs can form due to monoclonal expansion and those tumors can acquire inter-tumoral heterogeneity through branched evolution. Grade I and II meningiomas can occur in the same patient. Thus, the molecular make-up and clinical behavior of one tumor in MMs, cannot reliably lend insight into that of the others and suggests the clinical management strategy for MMs should be tailored individually.
Keywords: Clonal formation; Genomics; Sporadic multiple meningiomas.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

