A TetR family transcriptional regulator, SP_2854 can affect the butenyl-spinosyn biosynthesis by regulating glucose metabolism in Saccharopolyspora pogona
- PMID: 35568948
- PMCID: PMC9107242
- DOI: 10.1186/s12934-022-01808-2
A TetR family transcriptional regulator, SP_2854 can affect the butenyl-spinosyn biosynthesis by regulating glucose metabolism in Saccharopolyspora pogona
Abstract
Background: Butenyl-spinosyn produced by Saccharopolyspora pogona exhibits strong insecticidal activity and a broad pesticidal spectrum. Currently, important functional genes involve in butenyl-spinosyn biosynthesis remain unknown, which leads to difficulty in efficiently understanding its regulatory mechanism, and improving its production by metabolic engineering.
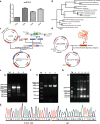
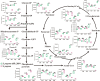
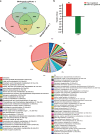
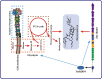
Results: Here, we identified a TetR family transcriptional regulator, SP_2854, that can positively regulate butenyl-spinosyn biosynthesis and affect strain growth, glucose consumption, and mycelial morphology in S. pogona. Using targeted metabolomic analyses, we found that SP_2854 overexpression enhanced glucose metabolism, while SP_2854 deletion had the opposite effect. To decipher the overproduction mechanism in detail, comparative proteomic analysis was carried out in the SP-2854 overexpressing mutant and the original strain, and we found that SP_2854 overexpression promoted the expression of proteins involved in glucose metabolism.
Conclusion: Our findings suggest that SP_2854 can affect strain growth and development and butenyl-spinosyn biosynthesis in S. pogona by controlling glucose metabolism. The strategy reported here will be valuable in paving the way for genetic engineering of regulatory elements in actinomycetes to improve important natural products production.
Keywords: Butenyl-spinosyn; Metabolic engineering; Omics analysis; SP_2854; Saccharopolyspora pogona.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Identification of a TetR family regulator and a polyketide synthase gene cluster involved in growth development and butenyl-spinosyn biosynthesis of Saccharopolyspora pogona.Appl Microbiol Biotechnol. 2021 Feb;105(4):1519-1533. doi: 10.1007/s00253-021-11105-4. Epub 2021 Jan 23. Appl Microbiol Biotechnol. 2021. PMID: 33484320
-
Bacterioferritin: a key iron storage modulator that affects strain growth and butenyl-spinosyn biosynthesis in Saccharopolyspora pogona.Microb Cell Fact. 2021 Aug 14;20(1):157. doi: 10.1186/s12934-021-01651-x. Microb Cell Fact. 2021. PMID: 34391414 Free PMC article.
-
Comparative Proteomics Reveals the Effect of the Transcriptional Regulator Sp13016 on Butenyl-Spinosyn Biosynthesis in Saccharopolyspora pogona.J Agric Food Chem. 2021 Oct 27;69(42):12554-12565. doi: 10.1021/acs.jafc.1c03654. Epub 2021 Oct 16. J Agric Food Chem. 2021. PMID: 34657420
-
Recent advances in the biochemistry of spinosyns.Appl Microbiol Biotechnol. 2009 Feb;82(1):13-23. doi: 10.1007/s00253-008-1784-8. Epub 2008 Dec 10. Appl Microbiol Biotechnol. 2009. PMID: 19082588 Review.
-
Genes for the biosynthesis of spinosyns: applications for yield improvement in Saccharopolyspora spinosa.J Ind Microbiol Biotechnol. 2001 Dec;27(6):399-402. doi: 10.1038/sj.jim.7000180. J Ind Microbiol Biotechnol. 2001. PMID: 11774006 Review.
Cited by
-
Effect of Precursors and Their Regulators on the Biosynthesis of Antibiotics in Actinomycetes.Molecules. 2024 Mar 3;29(5):1132. doi: 10.3390/molecules29051132. Molecules. 2024. PMID: 38474644 Free PMC article. Review.
-
The PurR family transcriptional regulator promotes butenyl-spinosyn production in Saccharopolyspora pogona.Appl Microbiol Biotechnol. 2025 Jan 21;109(1):14. doi: 10.1007/s00253-024-13390-1. Appl Microbiol Biotechnol. 2025. PMID: 39836216 Free PMC article.
-
Addition of vegetable oil to enhance the biosynthesis of butenyl-spinosyn in a high production strain Saccharopolyspora pogona.PLoS One. 2025 Mar 26;20(3):e0319332. doi: 10.1371/journal.pone.0319332. eCollection 2025. PLoS One. 2025. PMID: 40138321 Free PMC article.
-
Enhanced Natamycin production in Streptomyces gilvosporeus through phosphate tolerance screening and transcriptome-based analysis of high-yielding mechanisms.Microb Cell Fact. 2025 Apr 2;24(1):79. doi: 10.1186/s12934-025-02696-y. Microb Cell Fact. 2025. PMID: 40176084 Free PMC article.
-
The LysR family transcriptional regulator ORF-L16 regulates spinosad biosynthesis in Saccharopolyspora spinosa.Synth Syst Biotechnol. 2024 May 10;9(4):609-617. doi: 10.1016/j.synbio.2024.05.001. eCollection 2024 Dec. Synth Syst Biotechnol. 2024. PMID: 38784197 Free PMC article.
References
-
- Paul L, Donald RH, Laura LK, Dennis OD, Jeffrey RG, Gary DC, Thomas W, Thomas CS, Pat MRE, Paul RG. Discovery of the butenyl-spinosyn insecticides: novel macrolides from the new bacterial strain Saccharopolyspora pogona. Bioorg. Med Chem. 2009;17:4185–4196.
-
- Huang KX, Xia LQ, Zhang YM, Ding XZ, Zahn AJ. Recent advances in the biochemistry of spinosyns. Appl Microbiol Biotechnol. 2009;82:12–23. doi: 10.1007/s00253-008-1784-8. - DOI
-
- Sheng Z, Chen K, Li X. Advances in the biosynthesis of spinosad—a review. Wei Sheng Wu Xue Bao. 2016;56:397–405. - PubMed
-
- Araujo RDS, Lopes MP, Barbosa WF, Gonçalves WG, Fernandes KM, Martins GF, Tavares MG. Spinosad-mediated effects on survival, overall group activity and the midgut of workers of Partamona helleri (Hymenoptera: Apidae) Ecotoxicol Environ Saf. 2019;175:148–154. doi: 10.1016/j.ecoenv.2019.03.050. - DOI - PubMed
-
- Mendonça TP, Davi de Aquino J, Junio da Silva W, Mendes DR, Campos CF, Vieira JS, Barbosa NP, Carvalho Naves MP, Olegário de E, Alves de Rezende AA, Spanó MA, Bonetti AM, Vieira Santos VS, Pereira BB, Resende de Morais C. Genotoxic and mutagenic assessment of spinosad using bioassays with Tradescantia pallida and Drosophila melanogaster. Chemosphere. 2019;222:503–510. doi: 10.1016/j.chemosphere.2019.01.182. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

