SynBioStrainFinder: A microbial strain database of manually curated CRISPR/Cas genetic manipulation system information for biomanufacturing
- PMID: 35568950
- PMCID: PMC9107733
- DOI: 10.1186/s12934-022-01813-5
SynBioStrainFinder: A microbial strain database of manually curated CRISPR/Cas genetic manipulation system information for biomanufacturing
Abstract
Background: Microbial strain information databases provide valuable data for microbial basic research and applications. However, they rarely contain information on the genetic operating system of microbial strains.
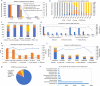
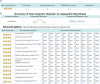
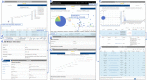
Results: We established a comprehensive microbial strain database, SynBioStrainFinder, by integrating CRISPR/Cas gene-editing system information with cultivation methods, genome sequence data, and compound-related information. It is presented through three modules, Strain2Gms/PredStrain2Gms, Strain2BasicInfo, and Strain2Compd, which combine to form a rapid strain information query system conveniently curated, integrated, and accessible on a single platform. To date, 1426 CRISPR/Cas gene-editing records of 157 microbial strains have been manually extracted from the literature in the Strain2Gms module. For strains without established CRISPR/Cas systems, the PredStrain2Gms module recommends the system of the most closely related strain as a reference to facilitate the construction of a new CRISPR/Cas gene-editing system. The database contains 139,499 records of strain cultivation and genome sequences, and 773,298 records of strain-related compounds. To facilitate simple and intuitive data application, all microbial strains are also labeled with stars based on the order and availability of strain information. SynBioStrainFinder provides a user-friendly interface for querying, browsing, and visualizing detailed information on microbial strains, and it is publicly available at http://design.rxnfinder.org/biosynstrain/ .
Conclusion: SynBioStrainFinder is the first microbial strain database with manually curated information on the strain CRISPR/Cas system as well as other microbial strain information. It also provides reference information for the construction of new CRISPR/Cas systems. SynBioStrainFinder will serve as a useful resource to extend microbial strain research and application for biomanufacturing.
Keywords: CRISPR/Cas system; Database; Gene-editing method; Genome sequence; Microorganism; Strain cultivation; Strain-related compound.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Wu L, Sun Q, Sugawara H, Yang S, Zhou Y, McCluskey K, Vasilenko A, Suzuki K, Ohkuma M, Lee Y, et al. Global catalogue of microorganisms (gcm): a comprehensive database and information retrieval, analysis, and visualization system for microbial resources. BMC Genomics. 2013;14:933. doi: 10.1186/1471-2164-14-933. - DOI - PMC - PubMed
MeSH terms
Grants and funding
- 2019YFA0904300/National Key Research and Development Program of China
- 31700081/National Natural Science Foundation of China
- 31570092/National Natural Science Foundation of China
- QYZDB-SSW-SMC012/CAS STS program
- 153D31KYSB20170121/International Partnership Program of Chinese Academy of Sciences of China
LinkOut - more resources
Full Text Sources

