Nontypeable Haemophilus influenzae infection of pulmonary macrophages drives neutrophilic inflammation in severe asthma
- PMID: 35570583
- PMCID: PMC9796932
- DOI: 10.1111/all.15375
Nontypeable Haemophilus influenzae infection of pulmonary macrophages drives neutrophilic inflammation in severe asthma
Abstract
Background: Nontypeable Haemophilus influenzae (NTHi) is a respiratory tract pathobiont that chronically colonizes the airways of asthma patients and is associated with severe, neutrophilic disease phenotypes. The mechanism of NTHi airway persistence is not well understood, but accumulating evidence suggests NTHi can persist within host airway immune cells such as macrophages. We hypothesized that NTHi infection of pulmonary macrophages drives neutrophilic inflammation in severe asthma.
Methods: Bronchoalveolar lavage (BAL) samples from 25 severe asthma patients were assessed by fluorescence in situ hybridisation to quantify NTHi presence. Weighted gene correlation network analysis (WGCNA) was performed on RNASeq data from NTHi-infected monocyte-derived macrophages to identify transcriptomic networks associated with NTHi infection.
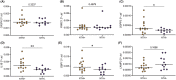
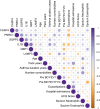
Results: NTHi was detected in 56% of BAL samples (NTHi+) and was associated with longer asthma duration (34 vs 22.5 years, p = .0436) and higher sputum neutrophil proportion (67% vs 25%, p = .0462). WGCNA identified a transcriptomic network of immune-related macrophage genes significantly associated with NTHi infection, including upregulation of T17 inflammatory mediators and neutrophil chemoattractants IL1B, IL8, IL23 and CCL20 (all p < .05). Macrophage network genes SGPP2 (p = .0221), IL1B (p = .0014) and GBP1 (p = .0477) were more highly expressed in NTHi+ BAL and moderately correlated with asthma duration (IL1B; rho = 0.41, p = .041) and lower prebronchodilator FEV1/FVC% (GBP1; rho = -0.43, p = .046 and IL1B; rho = -0.42, p = .055).
Conclusions: NTHi persistence with pulmonary macrophages may contribute to chronic airway inflammation and T17 responses in severe asthma, which can lead to decreased lung function and reduced steroid responsiveness. Identifying therapeutic strategies to reduce the burden of NTHi in asthma could improve patient outcomes.
Keywords: NTHi; T17 responses; asthma; inflammation; macrophage; neutrophil.
© 2022 The Authors. Allergy published by European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
DC reports that he was a postdoctoral researcher on projects funded by Pfizer and GSK between April 2014 and October 2017. RJK received a nonpromotional grant (£35,000) from Novartis to assist in funding initial database support for the WATCH study. PH reports that he is an employee of GSK. TW reports grants and personal fees from AstraZeneca, personal fees and others from MMH, grants and personal fees from GSK, personal fees from BI, and grants and personal fees from Synairgen, outside the submitted work. KS reports grants from AstraZeneca, outside the submitted work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol. 2015;16:45‐56. - PubMed
-
- Ishmael FT. The inflammatory response in the pathogenesis of asthma. J Am Osteopath Assoc. 2011;111:S11‐S17. - PubMed
-
- Deckers J, Branco Madeira F, Hammad H. Innate immune cells in asthma. Trends Immunol. 2013;34:540‐547. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

