Establishment of a lncRNA-Based Prognostic Gene Signature Associated With Altered Immune Responses in HCC
- PMID: 35572559
- PMCID: PMC9097819
- DOI: 10.3389/fimmu.2022.880288
Establishment of a lncRNA-Based Prognostic Gene Signature Associated With Altered Immune Responses in HCC
Abstract
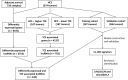
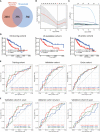
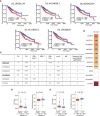
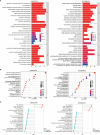
Hepatocellular carcinoma (HCC) is a common malignancy with higher mortality, and means are urgently needed to improve the prognosis. T cell exclusion (TCE) plays a pivotal role in immune evasion, and lncRNAs represent a large group of tumor development and progression modulators. Using the TCGA HCC dataset (n=374), we identified 2752 differentially expressed and 702 TCE-associated lncRNAs, of which 336 were in both groups. As identified using the univariate Cox regression analysis, those associated with overall survival (OS) were subjected to the LASSO-COX regression analysis to develop a prognosis signature. The model, which consisted of 11 lncRNAs and was named 11LNCPS for 11-lncRNA prognosis signature, was validated and performed better than two previous models. In addition to OS and TCE, higher 11LNCPS scores had a significant correlation with reduced infiltrations of CD8+ T cells and dendritic cells (DCs) and decreased infiltrations of Th1, Th2, and pro B cells. As expected, these infiltration alterations were significantly associated with worse OS in HCC. Analysis of published data indicates that HCCs with higher 11LNCPS scores were transcriptomically similar to those that responded better to PDL1 inhibitor. Of the 11LNCPS lncRNAs, LINC01134 and AC116025.2 seem more crucial, as their upregulations affected more immune cell types' infiltrations and were significantly associated with TCE, worse OS, and compromised immune responses in HCC. LncRNAs in the 11LNCPS impacted many cancer-associated biological processes and signaling pathways, particularly those involved in immune function and metabolism. The 11LNCPS should be useful for predicting prognosis and immune responses in HCC.
Keywords: AC116025.2; LINC01134; T cell exclusion; hepatocellular carcinoma (HCC); lncRNA; prognosis.
Copyright © 2022 Li, Zhang, Liu, Fu, A, Chen, Wu and Dong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Olson B, Li Y, Lin Y, Liu ET, Patnaik A. Mouse Models for Cancer Immunotherapy Research. Cancer Discov (2018) 8(11):1358–65. doi: 10.1158/2159-8290.CD-18-0044 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

